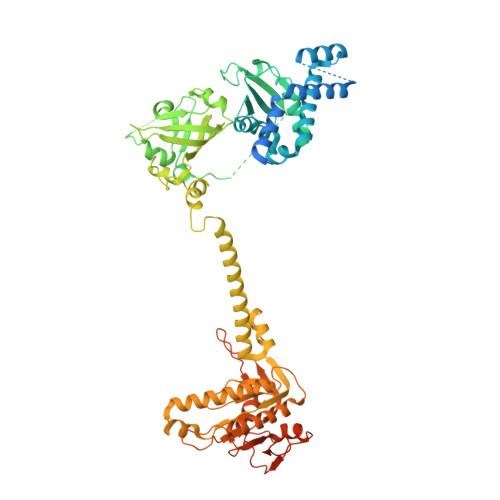
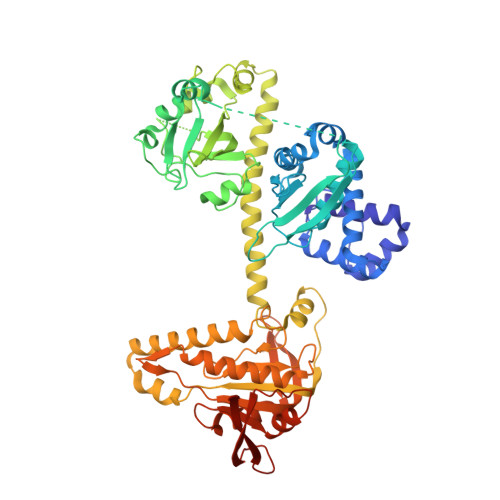
Structural insights into the mechanism of human soluble guanylate cyclase.
Kang, Y., Liu, R., Wu, J.X., Chen, L.(2019) Nature 574: 206-210
- PubMed: 31514202
- DOI: https://doi.org/10.1038/s41586-019-1584-6
- Primary Citation of Related Structures:
6JT0, 6JT1, 6JT2 - PubMed Abstract:
Soluble guanylate cyclase (sGC) is the primary sensor of nitric oxide. It has a central role in nitric oxide signalling and has been implicated in many essential physiological processes and disease conditions. The binding of nitric oxide boosts the enzymatic activity of sGC. However, the mechanism by which nitric oxide activates the enzyme is unclear. Here we report the cryo-electron microscopy structures of the human sGCα1β1 heterodimer in different functional states. These structures revealed that the transducer module bridges the nitric oxide sensor module and the catalytic module. Binding of nitric oxide to the β1 haem-nitric oxide and oxygen binding (H-NOX) domain triggers the structural rearrangement of the sensor module and a conformational switch of the transducer module from bending to straightening. The resulting movement of the N termini of the catalytic domains drives structural changes within the catalytic module, which in turn boost the enzymatic activity of sGC.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, Institute of Molecular Medicine, Peking University, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Beijing, China.