A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
Vangaveti, S., Cantara, W.A., Spears, J.L., DeMirci, H., Murphy IV, F.V., Ranganathan, S.V., Sarachan, K.L., Agris, P.F.(2020) J Mol Biology 432: 913-929
- PubMed: 31945376
- DOI: https://doi.org/10.1016/j.jmb.2019.12.016
- Primary Citation of Related Structures:
6DTI, 6MKN, 6MPF, 6MPI - PubMed Abstract:
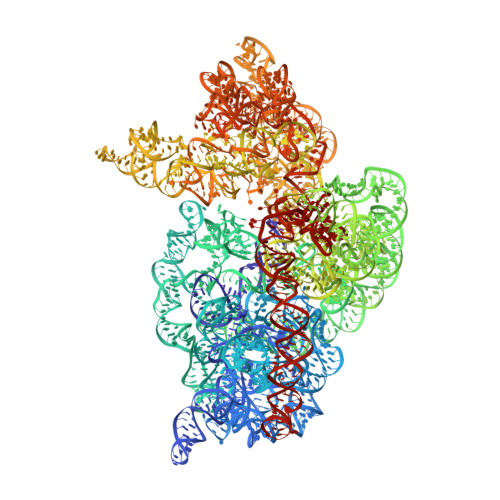
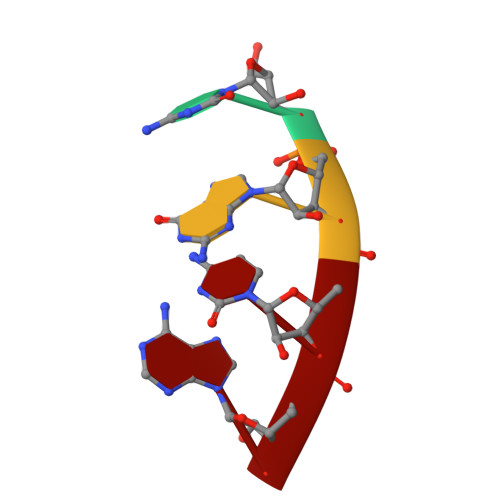
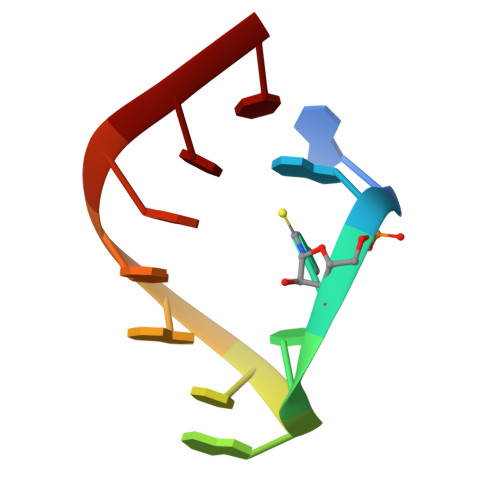
Three of six arginine codons (CGU, CGC, and CGA) are decoded by two Escherichia coli tRNA Arg isoacceptors. The anticodon stem and loop (ASL) domains of tRNA Arg1 and tRNA Arg2 both contain inosine and 2-methyladenosine modifications at positions 34 (I 34 ) and 37 (m 2 A 37 ). tRNA Arg1 is also modified from cytidine to 2-thiocytidine at position 32 (s 2 C 32 ). The s 2 C 32 modification is known to negate wobble codon recognition of the rare CGA codon by an unknown mechanism, while still allowing decoding of CGU and CGC. Substitution of s 2 C 32 for C 32 in the Saccharomyces cerevisiae tRNA Ile IAU anticodon stem and loop domain (ASL) negates wobble decoding of its synonymous A-ending codon, suggesting that this function of s 2 C at position 32 is a generalizable property. X-ray crystal structures of variously modified ASL Arg1 ICG and ASL Arg2 ICG constructs bound to cognate and wobble codons on the ribosome revealed the disruption of a C 32- A 38 cross-loop interaction but failed to fully explain the means by which s 2 C 32 restricts I 34 wobbling. Computational studies revealed that the adoption of a spatially broad inosine-adenosine base pair at the wobble position of the codon cannot be maintained simultaneously with the canonical ASL U-turn motif. C 32- A 38 cross-loop interactions are required for stability of the anticodon/codon interaction in the ribosomal A-site.
Organizational Affiliation:
The RNA Institute, University at Albany-SUNY, 1400 Washington Ave., Albany, NY, 12222, USA.