Architecture of the Cellulose Synthase Outer Membrane Channel and Its Association with the Periplasmic TPR Domain.
Acheson, J.F., Derewenda, Z.S., Zimmer, J.(2019) Structure 27: 1855
- PubMed: 31604608
- DOI: https://doi.org/10.1016/j.str.2019.09.008
- Primary Citation of Related Structures:
6TZK - PubMed Abstract:
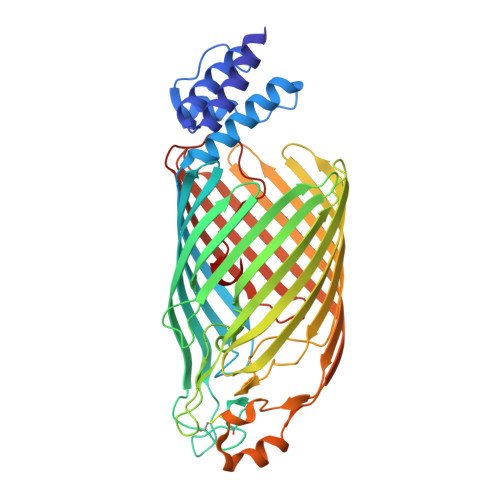
Extracellular bacterial cellulose contributes to biofilm stability and to the integrity of the bacterial cell envelope. In Gram-negative bacteria, cellulose is synthesized and secreted by a multi-component cellulose synthase complex. The BcsA subunit synthesizes cellulose and also transports the polymer across the inner membrane. Translocation across the outer membrane occurs through the BcsC porin, which extends into the periplasm via 19 tetra-tricopeptide repeats (TPR). We present the crystal structure of a truncated BcsC, encompassing the last TPR repeat and the complete outer membrane channel domain, revealing a 16-stranded, β barrel pore architecture. The pore is blocked by an extracellular gating loop, while the extended C terminus inserts deeply into the channel and positions a conserved Trp residue near its extracellular exit. The channel is lined with hydrophilic and aromatic residues suggesting a mechanism for facilitated cellulose diffusion based on aromatic stacking and hydrogen bonding.
- University of Virginia, School of Medicine, Department of Molecular Physiology and Biological Physics, Charlottesville, VA 22903, USA.
Organizational Affiliation: