Crystal Structure of Metallo Beta Lactamase from Hirschia baltica in the Complex with the Inhibitor Captopril.
Maltseva, N., Kim, Y., Clancy, S., Endres, M., Mulligan, R., Joachimiak, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Currently 6V61 does not have a validation slider image.
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-lactamase | 235 | Hirschia baltica ATCC 49814 | Mutation(s): 1 EC: 3.5.2.6 | 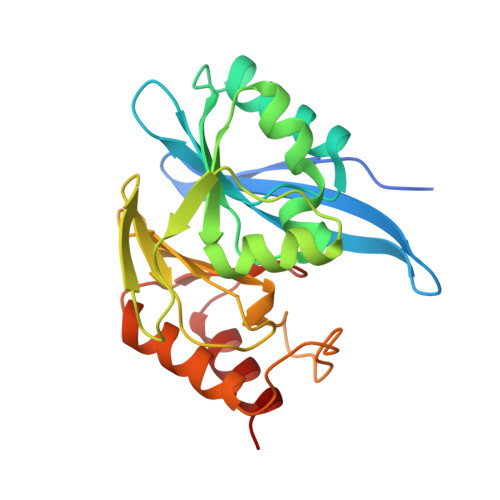 | |
UniProt | |||||
Find proteins for C6XID6 (Hirschia baltica (strain ATCC 49814 / DSM 5838 / IFAM 1418)) Explore C6XID6 Go to UniProtKB: C6XID6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C6XID6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| X8Z (Subject of Investigation/LOI) Query on X8Z | E [auth A] | L-CAPTOPRIL C9 H15 N O3 S FAKRSMQSSFJEIM-RQJHMYQMSA-N |  | ||
| ZN Query on ZN | B [auth A], C [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | D [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| FMT Query on FMT | H [auth A], I [auth A] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| CL Query on CL | J [auth A], K [auth A], L [auth A], M [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | F [auth A], G [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 77.893 | α = 90 |
| b = 77.893 | β = 90 |
| c = 241.426 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| HKL-3000 | phasing |
Currently 6V61 does not have a validation slider image.
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Human Genome Research Institute (NIH/NHGRI) | United States | -- |