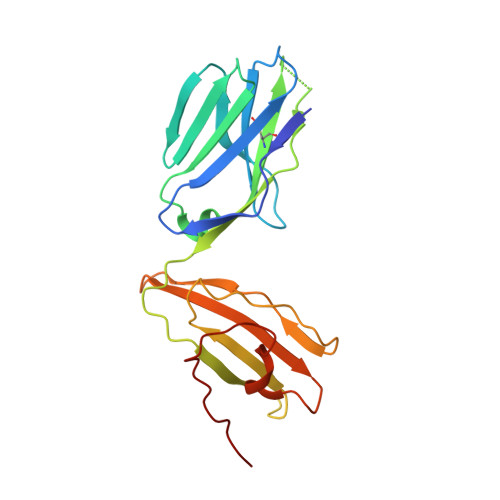
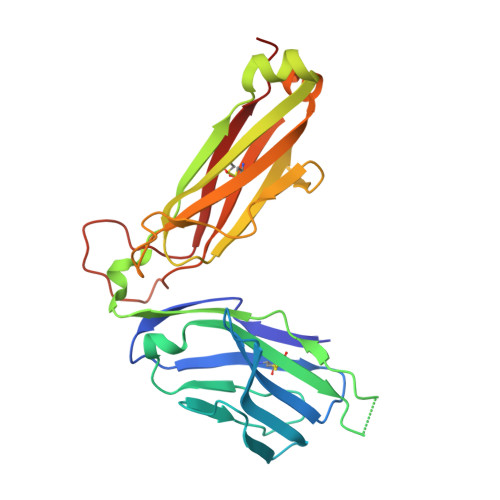
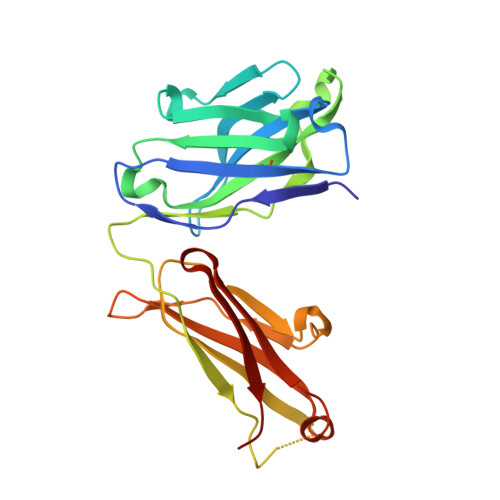
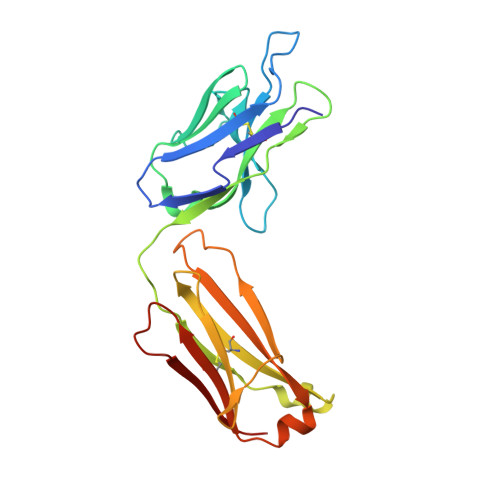
Structure-Guided Engineering of Immunotherapies Targeting TRBC1 and TRBC2 in T Cell Malignancies
Ferrari, M., Baldan, V., Wawrzyniecka, P., Bulek, A., Kinna, A., Ma, B., Bugda, R., Akbar, Z., Srivastava, S., Ghongane, P., Gannon, I., Robson, M., Sillibourne, J., Jha, R., Lim, W., Hopkins, J., Welin, M., Surade, S., Dyson, M., McCafferty, J., Cordoba, S., Thomas, S., Logan, D., Sewell, A., Maciocia, P., Onuoha, S., Pule, M.(2022) Res Sq