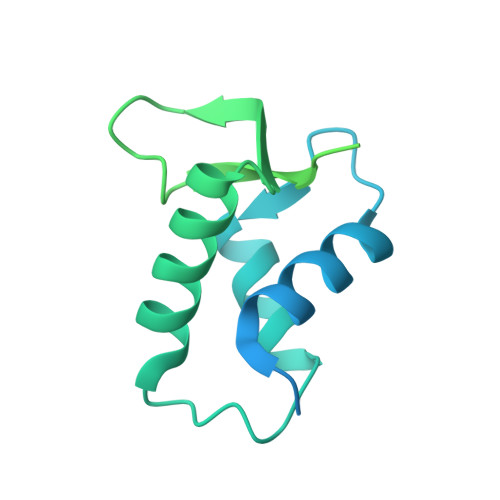
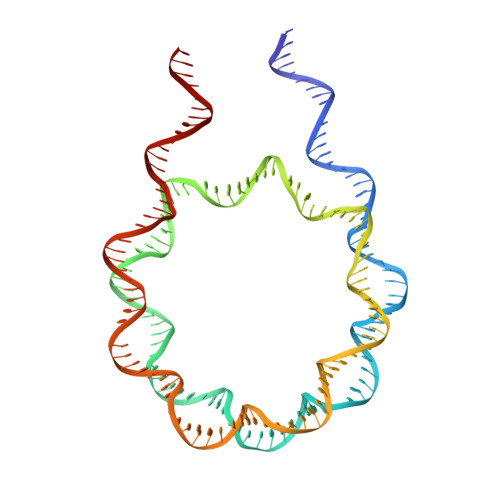
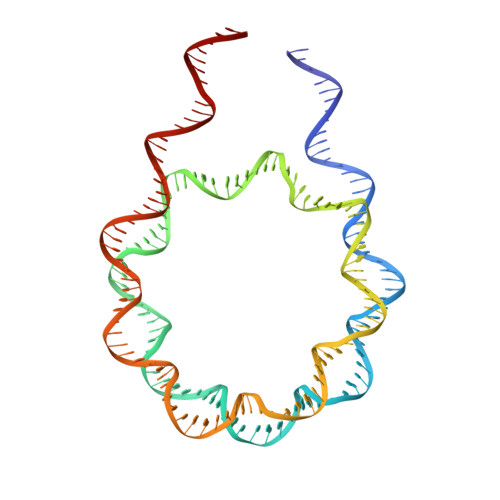
Structural features of nucleosomes in interphase and metaphase chromosomes.
Arimura, Y., Shih, R.M., Froom, R., Funabiki, H.(2021) Mol Cell 81: 4377
- PubMed: 34478647
- DOI: https://doi.org/10.1016/j.molcel.2021.08.010
- Primary Citation of Related Structures:
7KBD, 7KBE, 7KBF - PubMed Abstract:
Structural heterogeneity of nucleosomes in functional chromosomes is unknown. Here, we devise the template-, reference- and selection-free (TRSF) cryo-EM pipeline to simultaneously reconstruct cryo-EM structures of protein complexes from interphase or metaphase chromosomes. The reconstructed interphase and metaphase nucleosome structures are on average indistinguishable from canonical nucleosome structures, despite DNA sequence heterogeneity, cell-cycle-specific posttranslational modifications, and interacting proteins. Nucleosome structures determined by a decoy-classifying method and structure variability analyses reveal the nucleosome structural variations in linker DNA, histone tails, and nucleosome core particle configurations, suggesting that the opening of linker DNA, which is correlated with H2A C-terminal tail positioning, is suppressed in chromosomes. High-resolution (3.4-3.5 Å) nucleosome structures indicate DNA-sequence-independent stabilization of superhelical locations ±0-1 and ±3.5-4.5. The linker histone H1.8 preferentially binds to metaphase chromatin, from which chromatosome cryo-EM structures with H1.8 at the on-dyad position are reconstituted. This study presents the structural characteristics of nucleosomes in chromosomes.
- Laboratory of Chromosome and Cell Biology, The Rockefeller University, New York, NY 10065, USA. Electronic address: yarimura@rockefeller.edu.
Organizational Affiliation: