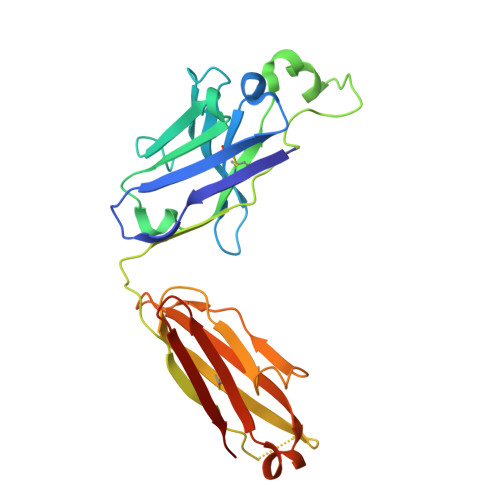
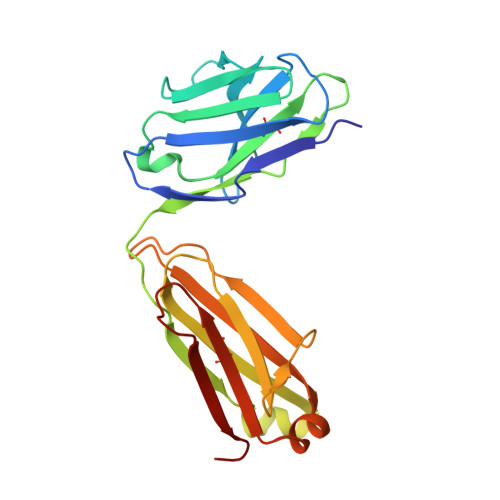
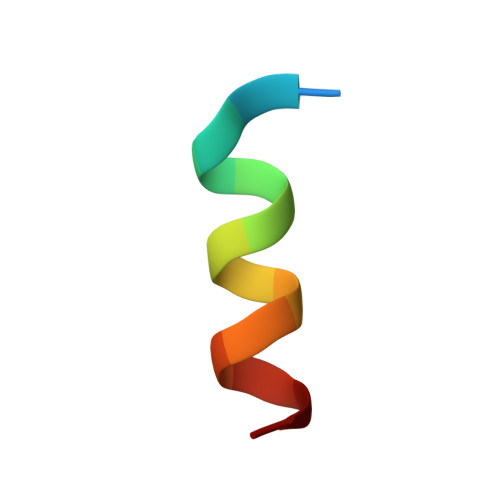
ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies.
Low, J.S., Jerak, J., Tortorici, M.A., McCallum, M., Pinto, D., Cassotta, A., Foglierini, M., Mele, F., Abdelnabi, R., Weynand, B., Noack, J., Montiel-Ruiz, M., Bianchi, S., Benigni, F., Sprugasci, N., Joshi, A., Bowen, J.E., Stewart, C., Rexhepaj, M., Walls, A.C., Jarrossay, D., Morone, D., Paparoditis, P., Garzoni, C., Ferrari, P., Ceschi, A., Neyts, J., Purcell, L.A., Snell, G., Corti, D., Lanzavecchia, A., Veesler, D., Sallusto, F.(2022) Science 377: 735-742
- PubMed: 35857703
- DOI: https://doi.org/10.1126/science.abq2679
- Primary Citation of Related Structures:
7SKZ, 7SL5, 7U09, 7U0A, 7U0E - PubMed Abstract:
The coronavirus spike glycoprotein attaches to host receptors and mediates viral fusion. Using a broad screening approach, we isolated seven monoclonal antibodies (mAbs) that bind to all human-infecting coronavirus spike proteins from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) immune donors. These mAbs recognize the fusion peptide and acquire affinity and breadth through somatic mutations. Despite targeting a conserved motif, only some mAbs show broad neutralizing activity in vitro against alpha- and betacoronaviruses, including animal coronaviruses WIV-1 and PDF-2180. Two selected mAbs also neutralize Omicron BA.1 and BA.2 authentic viruses and reduce viral burden and pathology in vivo. Structural and functional analyses showed that the fusion peptide-specific mAbs bound with different modalities to a cryptic epitope hidden in prefusion stabilized spike, which became exposed upon binding of angiotensin-converting enzyme 2 (ACE2) or ACE2-mimicking mAbs.
- Institute for Research in Biomedicine, Università della Svizzera Italiana, 6500 Bellinzona, Switzerland.
Organizational Affiliation: