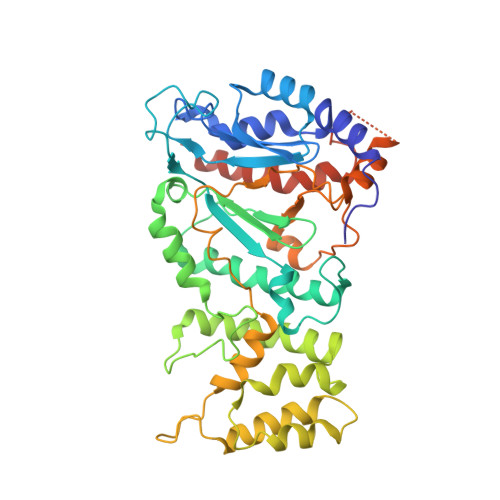
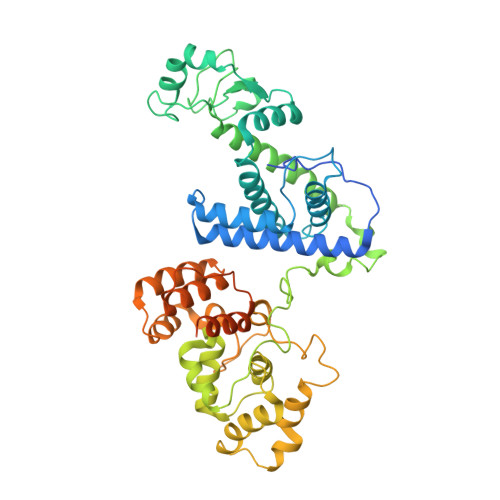
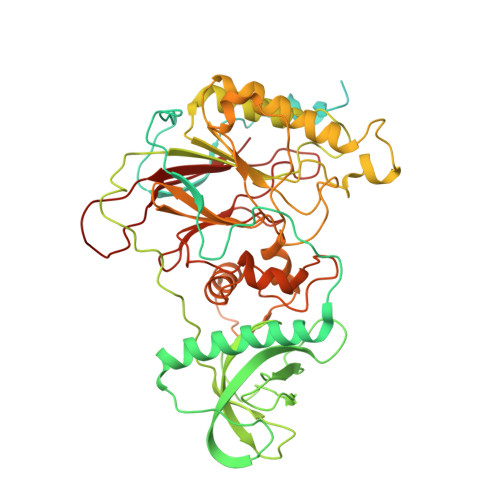
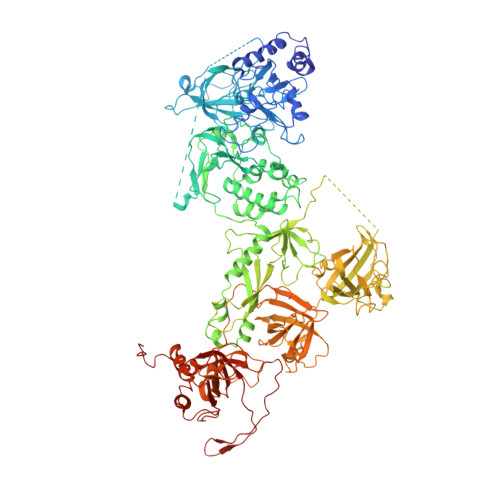
Cryo-EM structure of the human CST-Pol alpha /primase complex in a recruitment state.
Cai, S.W., Zinder, J.C., Svetlov, V., Bush, M.W., Nudler, E., Walz, T., de Lange, T.(2022) Nat Struct Mol Biol 29: 813-819
- PubMed: 35578024
- DOI: https://doi.org/10.1038/s41594-022-00766-y
- Primary Citation of Related Structures:
7U5C - PubMed Abstract:
The CST-Polα/primase complex is essential for telomere maintenance and functions to counteract resection at double-strand breaks. We report a 4.6-Å resolution cryo-EM structure of human CST-Polα/primase, captured prior to catalysis in a recruitment state stabilized by chemical cross-linking. Our structure reveals an evolutionarily conserved interaction between the C-terminal domain of the catalytic POLA1 subunit and an N-terminal expansion in metazoan CTC1. Cross-linking mass spectrometry and negative-stain EM analysis provide insight into CST binding by the flexible POLA1 N-terminus. Finally, Coats plus syndrome disease mutations previously characterized to disrupt formation of the CST-Polα/primase complex map to protein-protein interfaces observed in the recruitment state. Together, our results shed light on the architecture and stoichiometry of the metazoan fill-in machinery.
Organizational Affiliation:
Laboratory of Cell Biology and Genetics, The Rockefeller University, New York, NY, USA.