Cloning and structural basis of fluorescent protein color variants from identical species of sea anemone, Diadumene lineata.
Horiuchi, Y., Makabe, K., Laskaratou, D., Hatori, K., Sliwa, M., Mizuno, H., Hotta, J.I.(2023) Photochem Photobiol Sci 22: 1591-1601
- PubMed: 36943649
- DOI: https://doi.org/10.1007/s43630-023-00399-0
- Primary Citation of Related Structures:
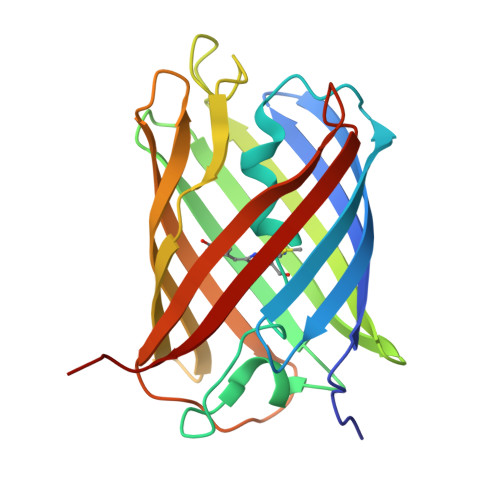
7X2B - PubMed Abstract:
Diadumene lineata is a colorful sea anemone with orange stripe tissue of the body column and plain tentacles with red lines. We subjected Diadumene lineata to expression cloning and obtained genes encoding orange (OFP: DiLiFP561) and red fluorescent proteins (RFPs: DiLiFP570 and DiLiFP571). These proteins formed obligatory tetramers. All three proteins showed bright fluorescence with the brightness of 58.3 mM -1 ·cm -1 (DiLiFP561), 43.9 mM -1 ·cm -1 (DiLiFP570), and 31.2 mM -1 ·cm -1 (DiLiFP571), which were equivalent to that of commonly used red fluorescent proteins. Amplitude-weighted average fluorescence lifetimes of DiLiFP561, DiLiFP570 and DiLiFP571 were determined as 3.7, 3.6 and 3.0 ns. We determined a crystal structure of DiLiFP570 at 1.63 Å resolution. The crystal structure of DiLiFP570 revealed that the chromophore has an extended π-conjugated structure similar to that of DsRed. Most of the amino acid residues surrounding the chromophore were common between DiLiFP570 and DiLiFP561, except M159 of DiLiFP570 (Lysine in DiLiFP561), which is located close to the chromophore hydroxyl group. Interestingly, a similar K-to-M substitution has been reported in a red-shifted variant of DsRed (mRFP1). It is a striking observation that the naturally evolved color-change variants are consistent with the mutation induced via protein engineering processes. The newly cloned proteins are promising as orange and red fluorescent markers for imaging with long fluorescence lifetime.
- Graduate School of Science and Engineering, Yamagata University, 4-3-16 Jonan, Yonezawa, Yamagata, 992-8510, Japan.
Organizational Affiliation: