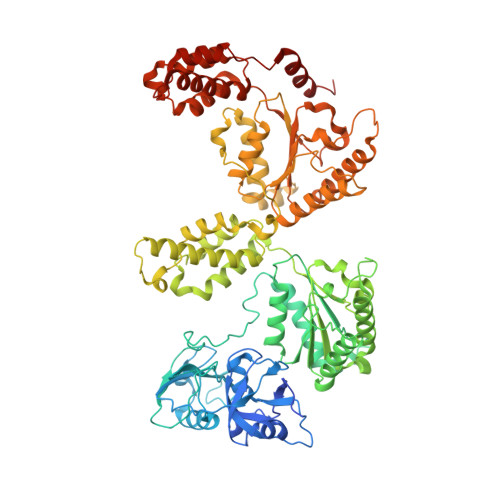
Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1.
Prattes, M., Grishkovskaya, I., Hodirnau, V.V., Hetzmannseder, C., Zisser, G., Sailer, C., Kargas, V., Loibl, M., Gerhalter, M., Kofler, L., Warren, A.J., Stengel, F., Haselbach, D., Bergler, H.(2022) Nat Struct Mol Biol 29: 942-953
- PubMed: 36097293
- DOI: https://doi.org/10.1038/s41594-022-00832-5
- Primary Citation of Related Structures:
7Z11, 7Z34 - PubMed Abstract:
The AAA-ATPase Drg1 is a key factor in eukaryotic ribosome biogenesis that initiates cytoplasmic maturation of the large ribosomal subunit. Drg1 releases the shuttling maturation factor Rlp24 from pre-60S particles shortly after nuclear export, a strict requirement for downstream maturation. The molecular mechanism of release remained elusive. Here, we report a series of cryo-EM structures that captured the extraction of Rlp24 from pre-60S particles by Saccharomyces cerevisiae Drg1. These structures reveal that Arx1 and the eukaryote-specific rRNA expansion segment ES27 form a joint docking platform that positions Drg1 for efficient extraction of Rlp24 from the pre-ribosome. The tips of the Drg1 N domains thereby guide the Rlp24 C terminus into the central pore of the Drg1 hexamer, enabling extraction by a hand-over-hand translocation mechanism. Our results uncover substrate recognition and processing by Drg1 step by step and provide a comprehensive mechanistic picture of the conserved modus operandi of AAA-ATPases.
Organizational Affiliation:
Institute of Molecular Biosciences, University of Graz, Graz, Austria.