Visualizing "Alternative Isoinformational Engineered" DNA in A- and B-Forms at High Resolution.
Hoshika, S., Shukla, M.S., Benner, S.A., Georgiadis, M.M.(2022) J Am Chem Soc 144: 15603-15611
- PubMed: 35969672
- DOI: https://doi.org/10.1021/jacs.2c05255
- Primary Citation of Related Structures:
8CRZ, 8CS0 - PubMed Abstract:
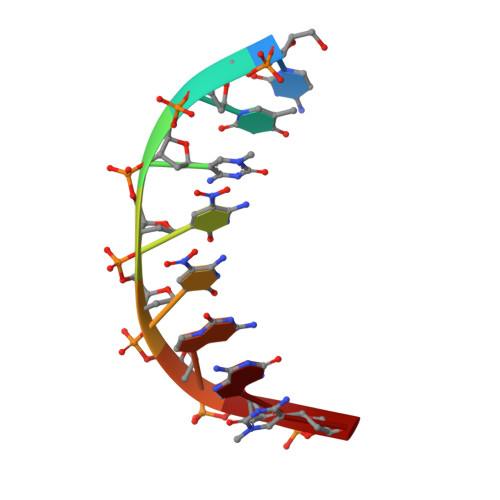
A fundamental property of DNA built from four informational nucleotide units (GCAT) is its ability to adopt different helical forms within the context of the Watson-Crick pair. Well-characterized examples include A-, B-, and Z-DNA. For this study, we created an isoinformational biomimetic polymer, built (like standard DNA) from four informational "letters", but with the building blocks being artificial. This ALternative Isoinformational ENgineered (ALIEN) DNA was hypothesized to support two nucleobase pairs, the P : Z pair matching 2-amino-imidazo-[1,2 a ]-1,3,5-triazin-[8 H ]-4-one with 6-amino-3-5-nitro-1 H -pyridin-2-one and the B : S pair matching 6-amino-4-hydroxy-5-1 H -purin-2-one with 3-methyl-6-amino-pyrimidin-2-one. We report two structures of ALIEN DNA duplexes at 1.2 Å resolution and a third at 1.65 Å. All of these are built from a single self-complementary sequence (5'-CT SZZPBSBSZPPB AG) that includes 12 consecutive ALIEN nucleotides. We characterized the helical, nucleobase pair, and dinucleotide step parameters of ALIEN DNA in these structures. In addition to showing that ALIEN pairs retain basic Watson-Crick pairing geometry, two of the ALIEN DNA structures are characterized as A-form DNA and one as B-form DNA. We identified parameters that map differences effecting the transition between the two helical forms; these same parameters distinguish helical forms of isoinformational natural DNA. Collectively, our analyses suggest that ALIEN DNA retains essential structural features of natural DNA, not only its information density and Watson-Crick pairing but also its ability to adopt two canonical forms.
- Foundation for Molecular Evolution, 13709 Progress Boulevard, No. 7, Alachua, Florida 32615, United States.
Organizational Affiliation: