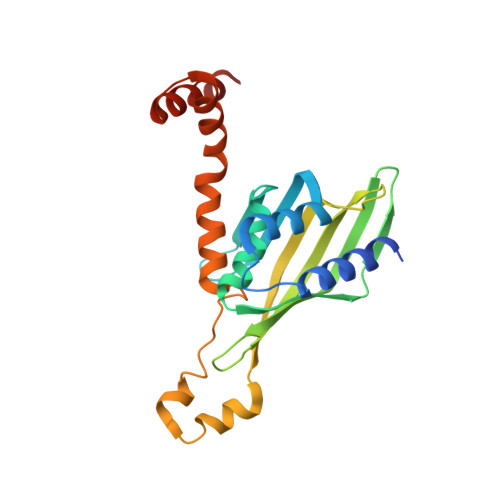
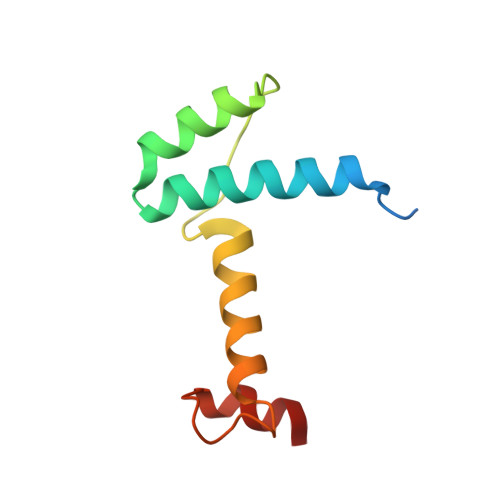
Structural basis of the interaction between BCL9-Pygo and LDB-SSBP complexes in assembling the Wnt enhanceosome.
Wang, H., Bienz, M., Yan, X.X., Xu, W.(2023) Nat Commun 14: 3702-3702
- PubMed: 37349336
- DOI: https://doi.org/10.1038/s41467-023-39439-9
- Primary Citation of Related Structures:
8HIB - PubMed Abstract:
The Wnt enhanceosome is responsible for transactivation of Wnt-responsive genes and a promising therapeutic target for treatment of numerous cancers with Adenomatous Polyposis Coli (APC) or β-catenin mutations. How the Wnt enhanceosome is assembled remains poorly understood. Here we show that B-cell lymphoma 9 protein (BCL9), Pygopus (Pygo), LIM domain-binding protein 1 (LDB1) and single-stranded DNA-binding protein (SSBP) form a stable core complex within the Wnt enhanceosome. Their mutual interactions rely on a highly conserved N-terminal asparagine proline phenylalanine (NPF) motif of Pygo, through which the BCL9-Pygo complex binds to the LDB-SSBP core complex. Our crystal structure of a ternary complex comprising the N-terminus of human Pygo2, LDB1 and SSBP2 reveals a single LDB1-SSBP2 complex binding simultaneously to two Pygo2 molecules via their NPF motifs. These interactions critically depend on the NPF motifs which bind to a deep groove formed between LDB1 and SSBP2, potentially constituting a binding site for drugs blocking Wnt/β-catenin signaling. Analysis of human cell lines lacking LDB or Pygo supports the functional relevance of the Pygo-LDB1-SSBP2 interaction for Wnt/β-catenin-dependent transcription.
- School of Life Science and Technology, ShanghaiTech University, Shanghai, China.
Organizational Affiliation: