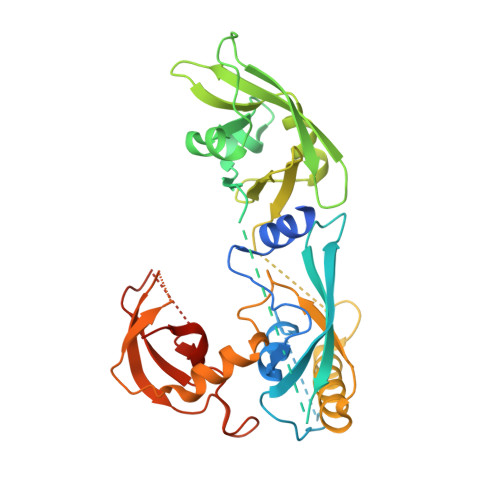
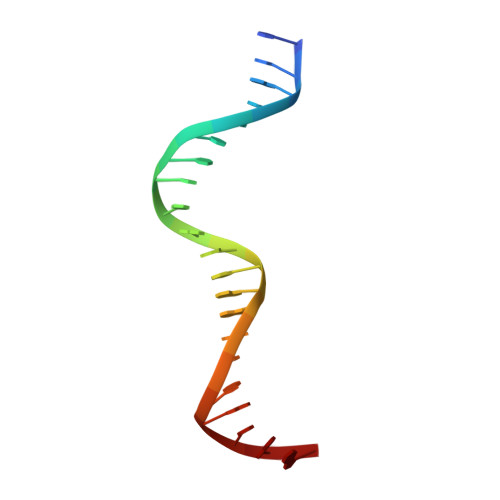
The structure and function of the DNA binding domain of class B MpARF2 share more traits with class A AtARF5 than to that of class B AtARF1.
Crespo, I., Malfois, M., Rienstra, J., Tarres-Sole, A., van den Berg, W., Weijers, D., Boer, D.R.(2025) Structure
- PubMed: 40086441
- DOI: https://doi.org/10.1016/j.str.2025.02.006
- Primary Citation of Related Structures:
8OJ1, 8OJ2 - PubMed Abstract:
Plant development is primarily controlled by the auxin phytohormones, which activate the auxin response factors (ARFs). Although the nuclear auxin pathway (NAP) is well studied, little is known on how ARFs specifically select target genes. Here, we investigated the DNA binding mechanism of ARF DNA binding domains (DBDs) from the activator class A and repressor class B in two evolutionary distant plant species, Marchantia polymorpha and Arabidopsis thaliana using fluorescence anisotropy, size exclusion chromatography, macromolecular crystallography (MX), and small-angle X-ray scattering (SAXS). We find that the previously proposed molecular caliper model, which partially explains the variability in binding of the ARFs to DNA, has been preserved throughout evolution. Our results show that the DBD of class B MpARF2 behaves more like class A AtARF5 than class B AtARF1. These findings suggest that DNA recognition of ARFs has diverged independently of the transcriptional output, which has significant implications for understanding diverse responses to auxin.
- ALBA Synchrotron Light Source, Carrer de la Llum 2-26, Cerdanyola del Vallès, 08290 Barcelona, Spain.
Organizational Affiliation: