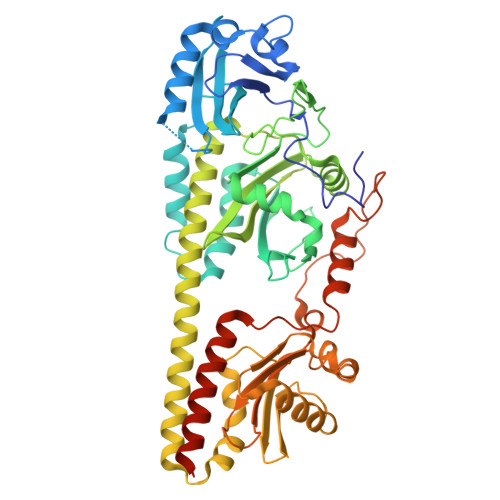
Serial-femtosecond crystallography reveals how a phytochrome variant couples chromophore and protein structural changes.
Sauthof, L., Szczepek, M., Schmidt, A., Bhowmick, A., Dasgupta, M., Mackintosh, M.J., Gul, S., Fuller, F.D., Chatterjee, R., Young, I.D., Michael, N., Heyder, N.A., Bauer, B., Koch, A., Bogacz, I., Kim, I.S., Simon, P.S., Butryn, A., Aller, P., Chukhutsina, V.U., Baxter, J.M., Hutchison, C.D.M., Liebschner, D., Poon, B., Sauter, N.K., Miller, M.D., Alonso-Mori, R., Hunter, M.S., Batyuk, A., Owada, S., Tono, K., Tanaka, R., van Thor, J.J., Krauss, N., Lamparter, T., Brewster, A.S., Schapiro, I., Orville, A.M., Yachandra, V.K., Yano, J., Hildebrandt, P., Kern, J.F., Scheerer, P.(2025) Sci Adv 11: eadp2665-eadp2665
- PubMed: 40435264
- DOI: https://doi.org/10.1126/sciadv.adp2665
- Primary Citation of Related Structures:
8RJM, 8RJN, 8RJO, 8RJP, 8RJQ, 8RJR, 8RJS, 8RJT, 8RJU - PubMed Abstract:
The photoreaction and commensurate structural changes of a chromophore within biological photoreceptors elicit conformational transitions of the protein promoting the switch between deactivated and activated states. We investigated how this coupling is achieved in a bacterial phytochrome variant, Agp2-PAiRFP2. Contrary to classical protein crystallography, which only allows probing (cryo-trapped) stable states, we have used time-resolved serial femtosecond x-ray crystallography (tr-SFX) and pump-probe techniques with various illumination and delay times with respect to photoexcitation of the parent Pfr state. Thus, structural data for seven time frames were sorted into groups of molecular events along the reaction coordinate. They range from chromophore isomerization to the formation of Meta-F, the intermediate that precedes the functional relevant secondary structure transition of the tongue. Structural data for the early events were used to calculate the photoisomerization pathway to complement the experimental data. Late events allow identifying the molecular switch that is linked to the intramolecular proton transfer as a prerequisite for the following structural transitions.
Organizational Affiliation:
Charité-Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Institute of Medical Physics and Biophysics, Group Structural Biology of Cellular Signaling, Charitéplatz 1, D-10117, Berlin, Germany.