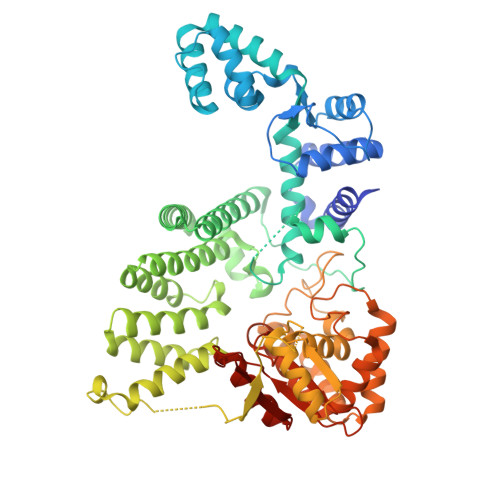
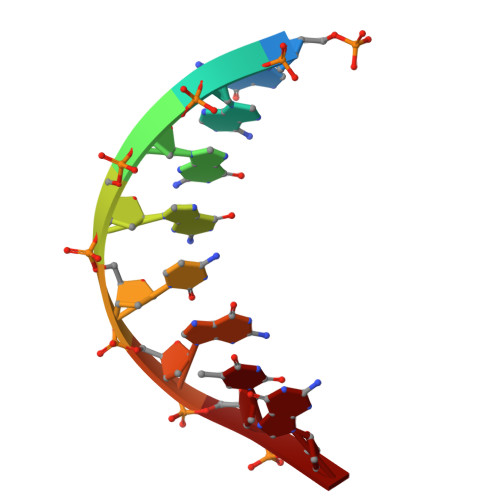
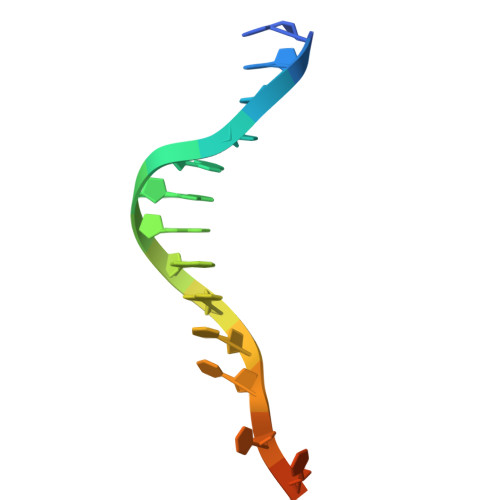
A FAN1 point mutation associated with accelerated Huntington's disease progression alters its PCNA-mediated assembly on DNA.
Aretz, J., Jeyasankar, G., Salerno-Kochan, A., Thomsen, M., Thieulin-Pardo, G., Haque, T., Monteagudo, E., Felsenfeld, D., Finley, M., Vogt, T.F., Boudet, J., Prasad, B.C.(2025) Nat Commun 16: 4412-4412
- PubMed: 40368883
- DOI: https://doi.org/10.1038/s41467-025-59324-x
- Primary Citation of Related Structures:
8S5A, 9EO1, 9EOA, 9GY0 - PubMed Abstract:
FAN1 is an endo- and exo-nuclease involved in DNA and interstrand crosslink repair. Genome-wide association studies of people with Huntington's disease revealed a strong association between the FAN1 R507H mutation and early disease onset, however the underlying mechanism(s) remains unclear. FAN1 has previously been implicated in modulating triplet repeat expansion in a PCNA dependent manner. To examine the role of PCNA on FAN1 activation, we solved the cryo-EM structures of a PCNA-FAN1-DNA complex. Our findings reveal that the FAN1 R507 residue directly interacts with PCNA D232. Biophysical interaction studies demonstrated that FAN1 enhances the binding affinity of PCNA for DNA, a synergistic effect disrupted in mutants carrying the R507H mutation. In contrast, PCNA does not affect the affinity of FAN1 for DNA but does modulate FAN1 activity upon ternary complex formation. The weakened and functionally altered FAN1 R507H-PCNA-DNA complex may partly impair the FAN1-mediated repair of CAG extrahelical extrusions, providing a potential explanation for the mutation's role in accelerating disease progression.
Organizational Affiliation:
Proteros biostructures GmbH, Bunsenstr. 7a, D - 82152, Martinsried, Germany.