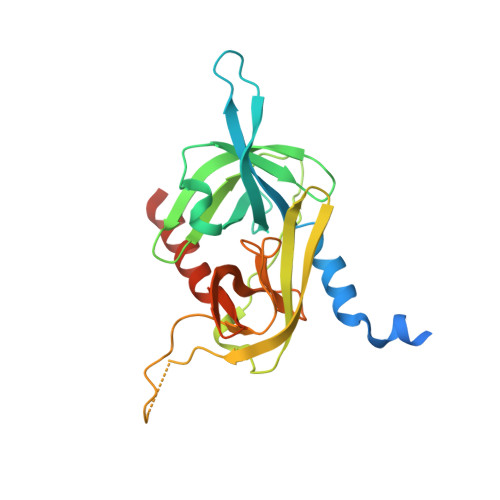
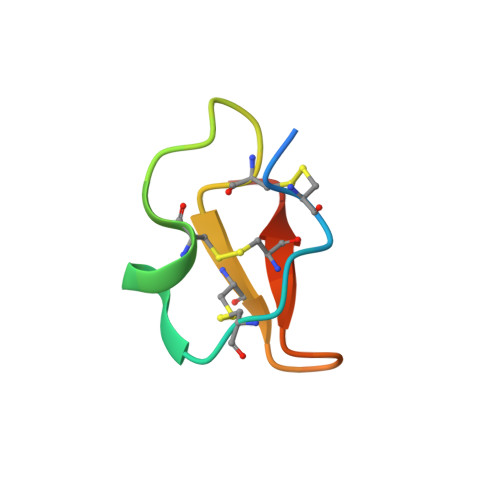
Cystine-knot peptide inhibitors of HTRA1 bind to a cryptic pocket within the active site region.
Li, Y., Wei, Y., Ultsch, M., Li, W., Tang, W., Tombling, B., Gao, X., Dimitrova, Y., Gampe, C., Fuhrmann, J., Zhang, Y., Hannoush, R.N., Kirchhofer, D.(2024) Nat Commun 15: 4359-4359
- PubMed: 38777835
- DOI: https://doi.org/10.1038/s41467-024-48655-w
- Primary Citation of Related Structures:
8SDM, 8SDP, 8SE7, 8SE8 - PubMed Abstract:
Cystine-knot peptides (CKPs) are naturally occurring peptides that exhibit exceptional chemical and proteolytic stability. We leveraged the CKP carboxypeptidase A1 inhibitor as a scaffold to construct phage-displayed CKP libraries and subsequently screened these collections against HTRA1, a trimeric serine protease implicated in age-related macular degeneration and osteoarthritis. The initial hits were optimized by using affinity maturation strategies to yield highly selective and potent picomolar inhibitors of HTRA1. Crystal structures, coupled with biochemical studies, reveal that the CKPs do not interact in a substrate-like manner but bind to a cryptic pocket at the S1' site region of HTRA1 and abolish catalysis by stabilizing a non-competent active site conformation. The opening and closing of this cryptic pocket is controlled by the gatekeeper residue V221, and its movement is facilitated by the absence of a constraining disulfide bond that is typically present in trypsin fold serine proteases, thereby explaining the remarkable selectivity of the CKPs. Our findings reveal an intriguing mechanism for modulating the activity of HTRA1, and highlight the utility of CKP-based phage display platforms in uncovering potent and selective inhibitors against challenging therapeutic targets.
- Department of Early Discovery Biochemistry, Genentech Inc., 1 DNA Way, South San Francisco, CA, 94080, USA.
Organizational Affiliation: