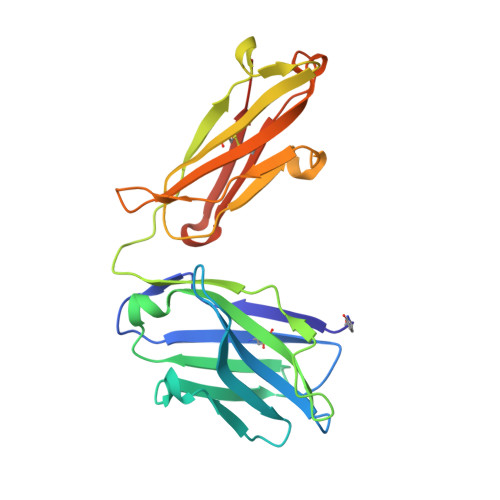
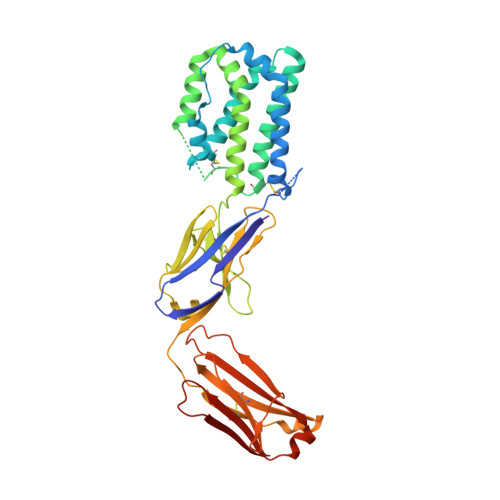
A novel interleukin-10 antibody graft to treat inflammatory bowel disease.
DiDonato, M., Simpson, C.T., Vo, T., Knuth, M., Geierstanger, B., Jamontt, J., Jones, D.H., Fathman, J.W., DeLarosa, D., Junt, T., Picard, D., Sommer, U., Bagger, M., Peters, E., Meeusen, S., Spraggon, G.(2025) Structure 33: 475-488.e7
- PubMed: 39798572
- DOI: https://doi.org/10.1016/j.str.2024.12.010
- Primary Citation of Related Structures:
8SVE - PubMed Abstract:
Inflammatory bowel disease (IBD) consists of chronic conditions that severely impact a patient's health and quality of life. Interleukin-10 (IL-10), a potent anti-inflammatory cytokine has strong genetic links to IBD susceptibility and has shown strong efficacy in IBD rodent models, suggesting it has great therapeutic potential. However, when tested in clinical trials for IBD, recombinant human IL-10 (rhIL-10) showed weak and inconsistent efficacy due to its short half-life and pro-inflammatory properties that counteract the anti-inflammatory efficacy. Here we present an engineered, IL-10, antibody-graft therapeutic (GFT-IL10M) designed to rectify these issues. GFT-IL10M combines the half-life extension properties of a monoclonal IgG antibody with altered IL-10 cell-type selective signaling, retaining desirable signaling on monocytes while reducing unwanted signaling on T, natural killer (NK), and B cells. Our structural and biochemical results indicate that the altered IL-10 topology in GFT-IL10M leads to a predominantly anti-inflammatory profile, potentially altering cell-type specific signaling patterns and extending half-life.
Organizational Affiliation:
Novartis Biomedical Research, 10675 John Jay Hopkins Drive, San Diego, CA 92121, USA.