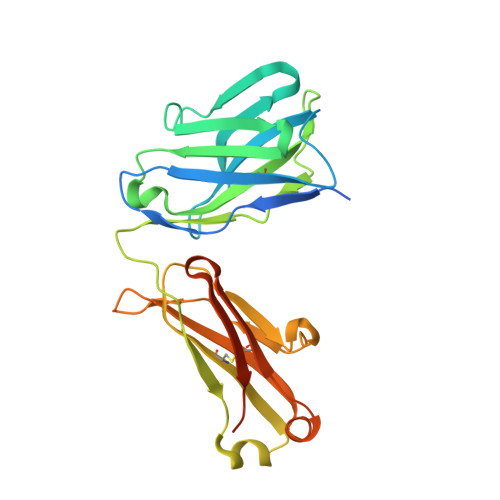
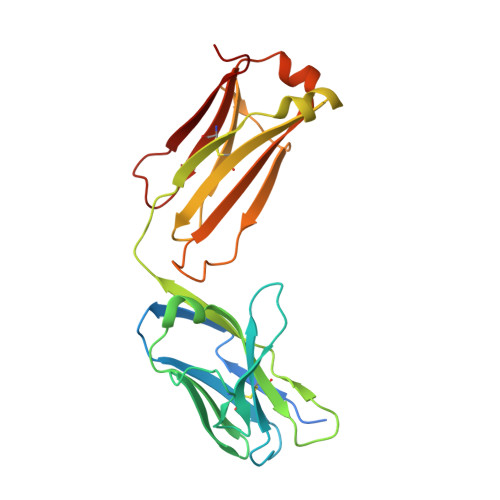
Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers.
Shahid, S., Karade, S.S., Hasan, S.S., Yin, R., Jiang, L., Liu, Y., Felbinger, N., Kulakova, L., Toth, E.A., Keck, Z.Y., Foung, S.K.H., Fuerst, T.R., Pierce, B.G., Mariuzza, R.A.(2025) Commun Biol 8: 825-825
- PubMed: 40442315
- DOI: https://doi.org/10.1038/s42003-025-08239-w
- Primary Citation of Related Structures:
8TFE, 8TGV, 8TGZ, 8THZ, 8TXQ, 8TZY, 8U9Y - PubMed Abstract:
Global elimination of hepatitis C virus (HCV) will require an effective cross-genotype vaccine. The HCV E2 envelope glycoprotein is the main target of neutralizing antibodies but also contains epitopes that elicit non-neutralizing antibodies which may provide protection through Fc effector functions rather than direct neutralization. We determined cryo-EM structures of a broadly neutralizing antibody, a moderately neutralizing antibody, and a non-neutralizing antibody bound to E2 to resolutions of 3.8, 3.3, and 3.7 Å, respectively. Whereas the broadly neutralizing antibody targeted the front layer of E2 and the non-neutralizing antibody targeted the back layer, the moderately neutralizing antibody straddled both front and back layers, and thereby defined a new neutralizing epitope on E2. The small size of complexes between conventional (monovalent) Fabs and E2 (~110 kDa) presented a challenge for cryo-EM. Accordingly, we engineered bivalent versions of E2-specific Fabs that doubled the size of Fab-E2 complexes and conferred highly identifiable shapes to the complexes that facilitated particle selection and orientation for image processing. This study validates bivalent Fabs as new fiducial markers for cryo-EM analysis of small proteins such as HCV E2 and identifies a new target epitope for vaccine development.
- W.M. Keck Laboratory for Structural Biology, University of Maryland-Institute for Bioscience and Biotechnology Research, Rockville, MD, USA.
Organizational Affiliation: