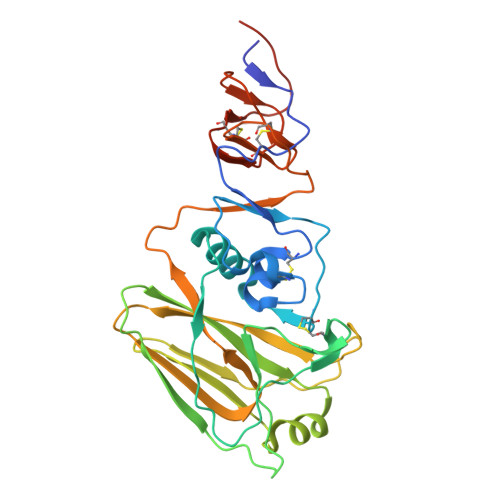
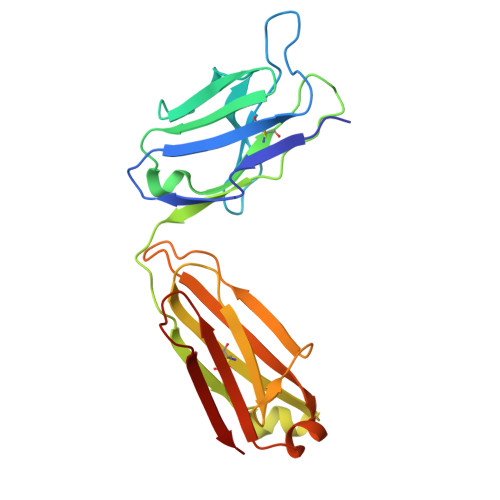
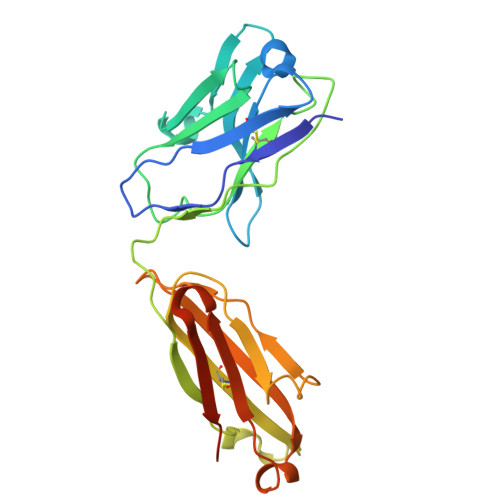
A protective and broadly binding antibody class engages the influenza virus hemagglutinin head at its stem interface.
Simmons, H.C., Finney, J., Kotaki, R., Adachi, Y., Park Moseman, A., Watanabe, A., Song, S., Robinson-McCarthy, L.R., Le Sage, V., Kuraoka, M., Moseman, E.A., Kelsoe, G., Takahashi, Y., McCarthy, K.R.(2024) bioRxiv
- PubMed: 38168412
- DOI: https://doi.org/10.1101/2023.12.13.571543
- Primary Citation of Related Structures:
8US0 - PubMed Abstract:
Influenza infection and vaccination impart strain-specific immunity that protects against neither seasonal antigenic variants nor the next pandemic. However, antibodies directed to conserved sites can confer broad protection. Here we identify and characterize a class of human antibodies that engage a previously undescribed, conserved epitope on the influenza hemagglutinin (HA) protein. Prototype antibody S8V1-157 binds at the normally occluded interface between the HA head and stem. Antibodies to this HA head-stem interface epitope are non-neutralizing in vitro but protect against lethal influenza infection in mice. Antibody isotypes that direct clearance of infected cells enhance this protection. Head-stem interface antibodies bind to most influenza A serotypes and seasonal human variants, and are present at low frequencies in the memory B cell populations of multiple human donors. Vaccines designed to elicit these antibodies might contribute to "universal" influenza immunity.
- Center for Vaccine Research, University of Pittsburgh School of Medicine, Pittsburgh, PA, USA.
Organizational Affiliation: