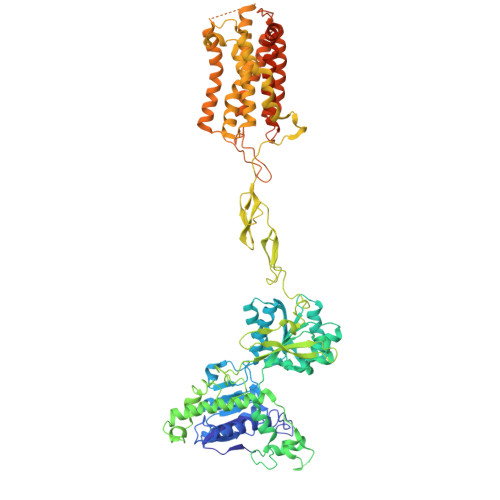
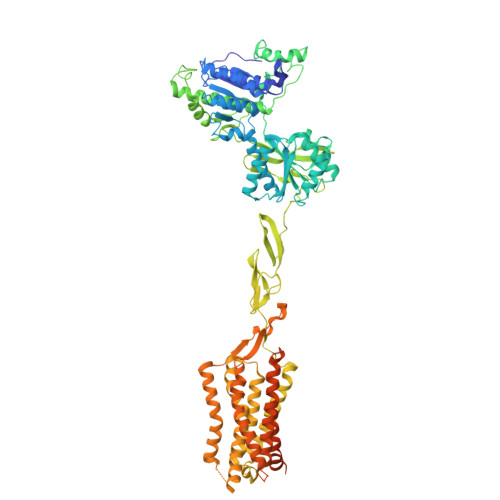
Structural basis of orientated asymmetry in a mGlu heterodimer.
Huang, W., Jin, N., Guo, J., Shen, C., Xu, C., Xi, K., Bonhomme, L., Quast, R.B., Shen, D.D., Qin, J., Liu, Y.R., Song, Y., Gao, Y., Margeat, E., Rondard, P., Pin, J.P., Zhang, Y., Liu, J.(2024) Nat Commun 15: 10345-10345
- PubMed: 39609406
- DOI: https://doi.org/10.1038/s41467-024-54744-7
- Primary Citation of Related Structures:
8WG9, 8WGB, 8WGC, 8WGD - PubMed Abstract:
The structural basis for the allosteric interactions within G protein-coupled receptors (GPCRs) heterodimers remains largely unknown. The metabotropic glutamate (mGlu) receptors are complex dimeric GPCRs important for the fine tuning of many synapses. Heterodimeric mGlu receptors with specific allosteric properties have been identified in the brain. Here we report four cryo-electron microscopy structures of mGlu2-4 heterodimer in different states: an inactive state bound to antagonists, two intermediate states bound to either mGlu2 or mGlu4 agonist only and an active state bound to both glutamate and a mGlu4 positive allosteric modulator (PAM) in complex with Gi protein. In addition to revealing a unique PAM binding pocket among mGlu receptors, our data bring important information for the asymmetric activation of mGlu heterodimers. First, we show that agonist binding to a single subunit in the extracellular domain is not sufficient to stabilize an active dimer conformation. Single-molecule FRET data show that the monoliganded mGlu2-4 can be found in both intermediate states and an active one. Second, we provide a detailed view of the asymmetric interface in seven-transmembrane (7TM) domains and identified key residues within the mGlu2 7TM that limits its activation leaving mGlu4 as the only subunit activating G proteins.
- Key Laboratory of Molecular Biophysics of MOE, College of Life Science and Technology, Huazhong University of Science and Technology (HUST), Wuhan, China.
Organizational Affiliation: