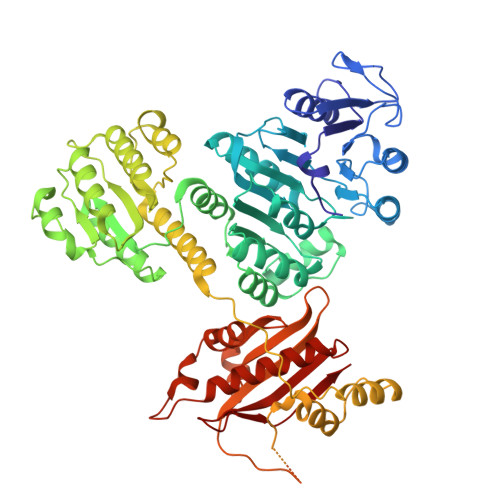
Crystal structure of the 4-hydroxybutyryl-CoA synthetase (ADP-forming) from nitrosopumilus maritimus.
Johnson, J., Tolar, B.B., Tosun, B., Yoshikuni, Y., Francis, C.A., Wakatsuki, S., DeMirci, H.(2024) Commun Biol 7: 1364-1364
- PubMed: 39433970
- DOI: https://doi.org/10.1038/s42003-024-06432-x
- Primary Citation of Related Structures:
8WZU - PubMed Abstract:
The 3-hydroxypropionate/4-hydroxybutyrate (3HP/4HB) cycle from ammonia-oxidizing Thaumarchaeota is currently considered the most energy-efficient aerobic carbon fixation pathway. The Nitrosopumilus maritimus 4-hydroxybutyryl-CoA synthetase (ADP-forming; Nmar_0206) represents one of several enzymes from this cycle that exhibit increased efficiency over crenarchaeal counterparts. This enzyme reduces energy requirements on the cell, reflecting thaumarchaeal success in adapting to low-nutrient environments. Here we show the structure of Nmar_0206 from Nitrosopumilus maritimus SCM1, which reveals a highly conserved interdomain linker loop between the CoA-binding and ATP-grasp domains. Phylogenetic analysis suggests the widespread prevalence of this loop and highlights both its underrepresentation within the PDB and structural importance within the (ATP-forming) acyl-CoA synthetase (ACD) superfamily. This linker is shown to have a possible influence on conserved interface interactions between domains, thereby influencing homodimer stability. These results provide a structural basis for the energy efficiency of this key enzyme in the modified 3HP/4HB cycle of Thaumarchaeota.
- Department of Molecular Biology and Genetics, Koç University, Istanbul, Türkiye.
Organizational Affiliation: