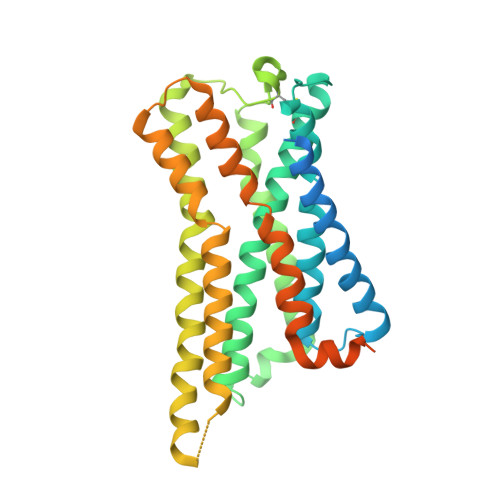
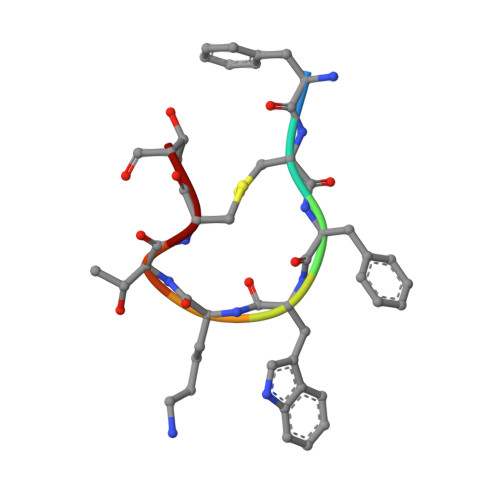
Structural basis for activation of somatostatin receptor 5 by cyclic neuropeptide agonists.
Li, J., You, C., Li, Y., Li, C., Fan, W., Chen, Z., Hu, W., Wu, K., Xu, H.E., Zhao, L.H.(2024) Proc Natl Acad Sci U S A 121: e2321710121-e2321710121
- PubMed: 38885377
- DOI: https://doi.org/10.1073/pnas.2321710121
- Primary Citation of Related Structures:
8X8L, 8X8N - PubMed Abstract:
Somatostatin receptor 5 (SSTR5) is an important G protein-coupled receptor and drug target for neuroendocrine tumors and pituitary disorders. This study presents two high-resolution cryogenicelectron microscope structures of the SSTR5-G i complexes bound to the cyclic neuropeptide agonists, cortistatin-17 (CST17) and octreotide, with resolutions of 2.7 Å and 2.9 Å, respectively. The structures reveal that binding of these peptides causes rearrangement of a "hydrophobic lock", consisting of residues from transmembrane helices TM3 and TM6. This rearrangement triggers outward movement of TM6, enabling Gα i protein engagement and receptor activation. In addition to hydrophobic interactions, CST17 forms conserved polar contacts similar to somatostatin-14 binding to SSTR2, while further structural and functional analysis shows that extracellular loops differently recognize CST17 and octreotide. These insights elucidate agonist selectivity and activation mechanisms of SSTR5, providing valuable guidance for structure-based drug development targeting this therapeutically relevant receptor.
- School of Chinese Materia Medica, Nanjing University of Chinese Medicine, Nanjing 210023, China.
Organizational Affiliation: