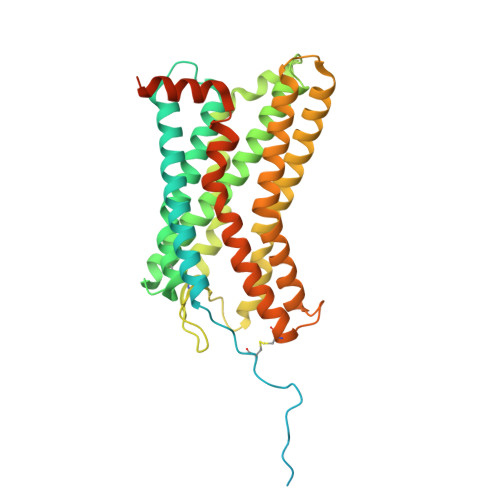
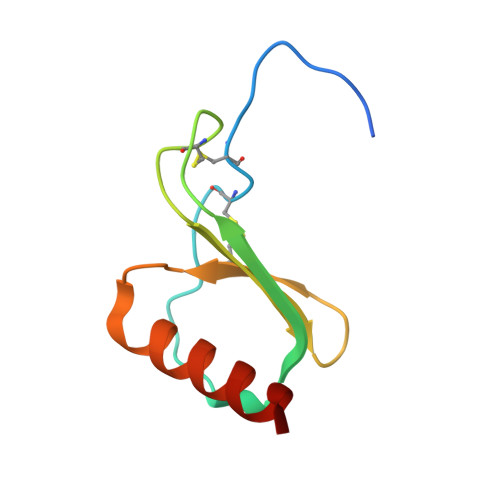
Molecular basis of promiscuous chemokine binding and structural mimicry at the C-X-C chemokine receptor, CXCR2.
Saha, S., Sano, F.K., Sharma, S., Ganguly, M., Mishra, S., Dalal, A., Akasaka, H., Kobayashi, T.A., Zaidi, N., Tiwari, D., Roy, N., Yadav, M.K., Banerjee, N., Saha, S., Mohapatra, S., Itoh, Y., Chevigne, A., Banerjee, R., Shihoya, W., Nureki, O., Shukla, A.K.(2025) Mol Cell 85: 976-988.e9
- PubMed: 39978339
- DOI: https://doi.org/10.1016/j.molcel.2025.01.024
- Primary Citation of Related Structures:
8XVU, 8XWA, 8XWF, 8XWM, 8XWN, 8XWS, 8XWV, 8XX3, 8XX6, 8XX7, 8XXH, 8XXR, 8XXX - PubMed Abstract:
Selectivity of natural agonists for their cognate receptors is a hallmark of G-protein-coupled receptors (GPCRs); however, this selectivity often breaks down at the chemokine receptors. Chemokines often display promiscuous binding to chemokine receptors, but the underlying molecular determinants remain mostly elusive. Here, we perform a comprehensive transducer-coupling analysis, testing all known C-X-C chemokines on every C-X-C type chemokine receptor to generate a global fingerprint of the selectivity and promiscuity encoded within this system. Taking lead from this, we determine cryoelectron microscopy (cryo-EM) structures of the most promiscuous receptor, C-X-C chemokine receptor 2 (CXCR2), in complex with several chemokines. These structural snapshots elucidate the details of ligand-receptor interactions, including structural motifs, which are validated using mutagenesis and functional experiments. We also observe that most chemokines position themselves on CXCR2 as a dimer while CXCL6 exhibits a monomeric binding pose. Taken together, our findings provide the molecular basis of chemokine promiscuity at CXCR2 with potential implications for developing therapeutic molecules.
- Department of Biological Sciences, Indian Institute of Technology Kanpur, Kanpur, India.
Organizational Affiliation: