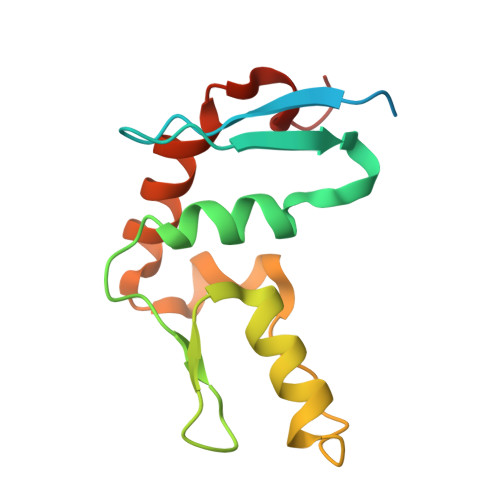
Structural basis for C. elegans pairing center DNA binding specificity by the ZIM/HIM-8 family proteins.
Li, M., Zhu, C., Xu, Z., Xu, M., Kuang, Y., Hou, X., Huang, X., Lv, M., Liu, Y., Zhang, Y., Xu, Z., Han, X., Wang, S., Shi, Y., Guang, S., Li, F.(2024) Nat Commun 15: 10355-10355
- PubMed: 39609407
- DOI: https://doi.org/10.1038/s41467-024-54548-9
- Primary Citation of Related Structures:
8YV9, 8YVA, 8YVB - PubMed Abstract:
Pairing center (PC) on each chromosome of Caenorhabditis elegans is crucial for homolog pairing and initiating synapsis. Within each PC, clusters of 11/12 bp DNA motif recruit one of four paralogous meiosis-specific proteins: ZIM-1, ZIM-2, ZIM-3, or HIM-8. However, the mechanistic basis underlying the specificity of ZIM/HIM-8-PC DNA interaction remains elusive. Here, we describe crystal structures of HIM-8, ZIM-1 and ZIM-2 DNA binding domains (ZF1, ZF2 and CTD) in complex with their cognate PC DNA motifs, respectively. These structures demonstrated the ZF1-2-CTD folds as an integrated structural unit crucial for its DNA binding specificity. Base-specific DNA-contacting residues are exclusively distributed on ZF1-2 and highly conserved. Furthermore, the CTD potentially contributes to the conformational diversity of ZF1-2, imparting binding specificity to distinct PC DNA motifs. These findings shed light on the mechanism governing PC DNA motif recognition by ZIM/HIM-8 proteins, suggesting a co-evolution relationship between PC DNA motifs and ZF1-2-CTD in shaping the specific recognition.
- MOE Key Laboratory for Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, The First Affiliated Hospital of USTC, Biomedical Sciences and Health Laboratory of Anhui Province, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China.
Organizational Affiliation: