Profiling influenza neuraminidase pre-existing humoral immunity in humans
Wang, X., Yang, Q., Chu, B.X., Wu, X.D., Zhang, Z.L.(2024) Cell Host Microbe
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2024) Cell Host Microbe
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 4N2C402_Fab_H | A, D [auth F], G [auth I], J [auth L] | 135 | Homo sapiens | Mutation(s): 0 | 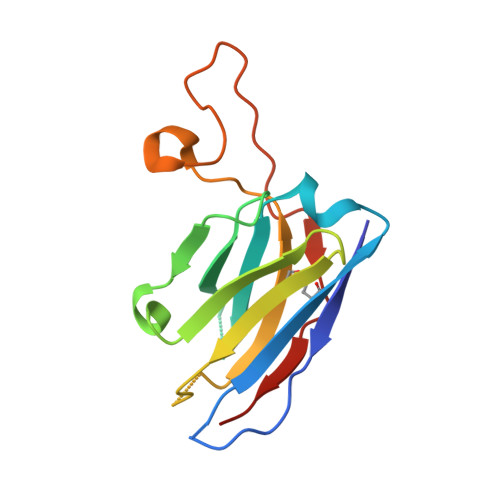 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 4N2C402_Fab_L | B, E [auth G], H [auth J], K [auth M] | 113 | Homo sapiens | Mutation(s): 0 | 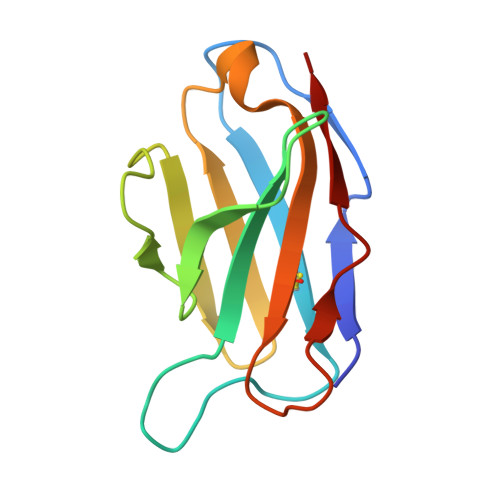 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Neuraminidase | C [auth D], F [auth H], I [auth K], L [auth N] | 393 | Influenza A virus | Mutation(s): 0 Gene Names: NA EC: 3.2.1.18 | 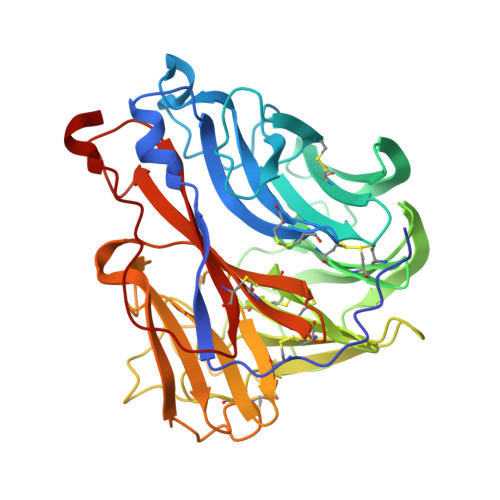 |
UniProt | |||||
Find proteins for A0A2S0X6Y5 (Influenza A virus) Explore A0A2S0X6Y5 Go to UniProtKB: A0A2S0X6Y5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2S0X6Y5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | M [auth O], N [auth P], O [auth Q], P [auth R] | 5 |  | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G22768VO GlyCosmos: G22768VO GlyGen: G22768VO | |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG (Subject of Investigation/LOI) Query on NAG | AA [auth N] Q [auth D] R [auth D] T [auth H] U [auth H] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| CA (Subject of Investigation/LOI) Query on CA | BA [auth N], S [auth D], V [auth H], Y [auth K] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other private | China | -- |