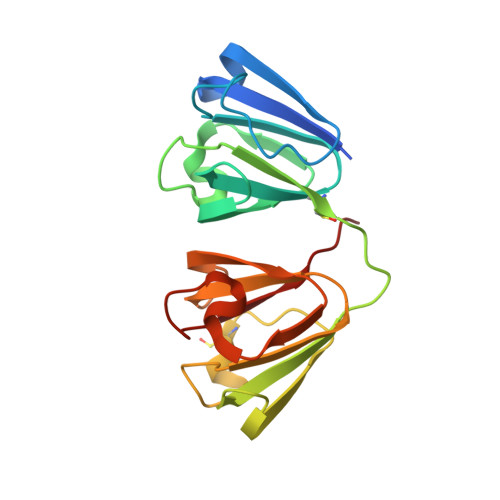
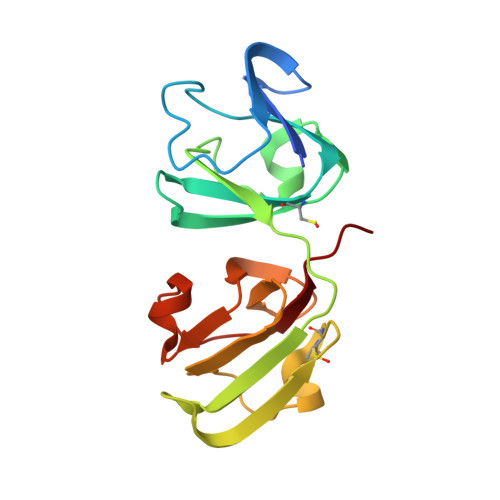
An ultraviolet-driven rescue pathway for oxidative stress to eye lens protein human gamma-D crystallin.
Hill, J.A., Nyathi, Y., Horrell, S., von Stetten, D., Axford, D., Owen, R.L., Beddard, G.S., Pearson, A.R., Ginn, H.M., Yorke, B.A.(2024) Commun Chem 7: 81-81
- PubMed: 38600176
- DOI: https://doi.org/10.1038/s42004-024-01163-w
- Primary Citation of Related Structures:
8BD0, 8BPI, 8Q3L - PubMed Abstract:
Human gamma-D crystallin (HGD) is a major constituent of the eye lens. Aggregation of HGD contributes to cataract formation, the leading cause of blindness worldwide. It is unique in its longevity, maintaining its folded and soluble state for 50-60 years. One outstanding question is the structural basis of this longevity despite oxidative aging and environmental stressors including ultraviolet radiation (UV). Here we present crystallographic structures evidencing a UV-induced crystallin redox switch mechanism. The room-temperature serial synchrotron crystallographic (SSX) structure of freshly prepared crystallin mutant (R36S) shows no post-translational modifications. After aging for nine months in the absence of light, a thiol-adduct (dithiothreitol) modifying surface cysteines is observed by low-dose SSX. This is shown to be UV-labile in an acutely light-exposed structure. This suggests a mechanism by which a major source of crystallin damage, UV, may also act as a rescuing factor in a finely balanced redox system.
- School of Chemistry and Biosciences, University of Bradford, Richmond Road, Bradford, BD7 1DP, United Kingdom.
Organizational Affiliation: