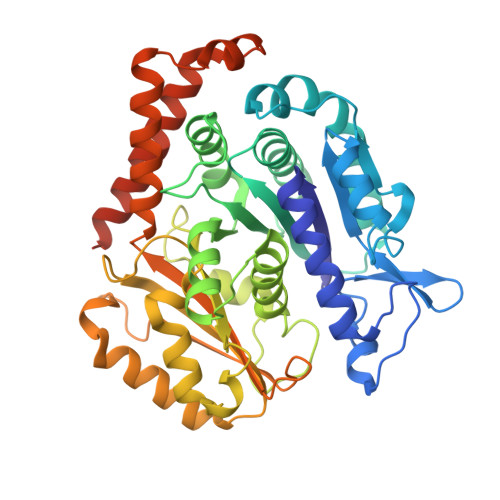
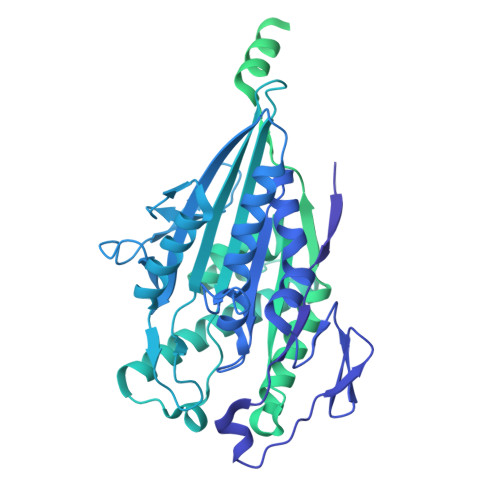
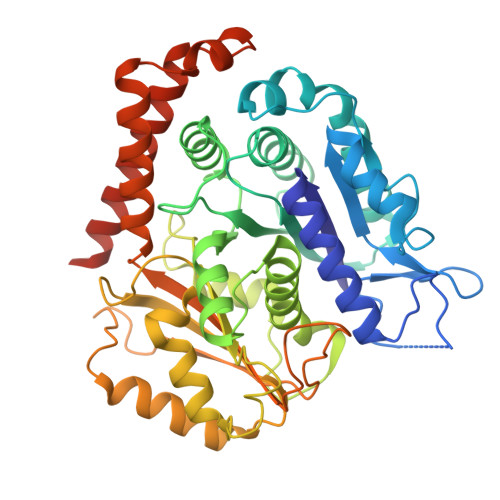
Microtubule association induces a Mg-free apo-like ADP pre-release conformation in kinesin-1 that is unaffected by its autoinhibitory tail.
Atherton, J., Chegkazi, M.S., Leusciatti, M., Di Palma, M., Peirano, E., Pozzer, L.S., Meli, M.V.A., Pasqualato, S., Foran, T., Morra, G., Steiner, R.A.(2025) Nat Commun 16: 6214-6214
- PubMed: 40617823
- DOI: https://doi.org/10.1038/s41467-025-61498-3
- Primary Citation of Related Structures:
8RHB, 8RHH, 8RIK, 8RIZ, 9GNQ - PubMed Abstract:
Kinesin-1 is a processive dimeric ATP-driven motor that transports vital intracellular cargos along microtubules (MTs). If not engaged in active transport, kinesin-1 limits futile ATP hydrolysis by adopting a compact autoinhibited conformation that involves an interaction between its C-terminal tail and the N-terminal motor domains. Here, using a chimeric kinesin-1 that fuses the N-terminal motor region to the tail and a tail variant unable to interact with the motors, we employ cryo-EM to investigate elements of the MT-associated mechanochemical cycle. We describe a missing structure for the proposed two-step allosteric mechanism of ADP release, the ATPase rate limiting step. It shows that MT association remodels the hydrogen bond network at the nucleotide binding site triggering removal of the Mg 2+ ion from the Mg 2+ -ADP complex. This results in a strong MT-binding apo-like state before ADP dissociation, which molecular dynamics simulations indicate is mediated by loop 9 dynamics. We further demonstrate that tail association does not directly affect this mechanism, nor the adoption of the ATP hydrolysis-competent conformation, nor neck linker docking/undocking, even when zippering the two motor domains. We propose a revised mechanism for tail-dependent kinesin-1 autoinhibition and suggest a possible explanation for its characteristic pausing behavior on MTs.
- Randall Centre for Cell and Molecular Biophysics, King's College London - New Hunt's House, Guy's Campus, London, UK. joseph.atherton@kcl.ac.uk.
Organizational Affiliation: