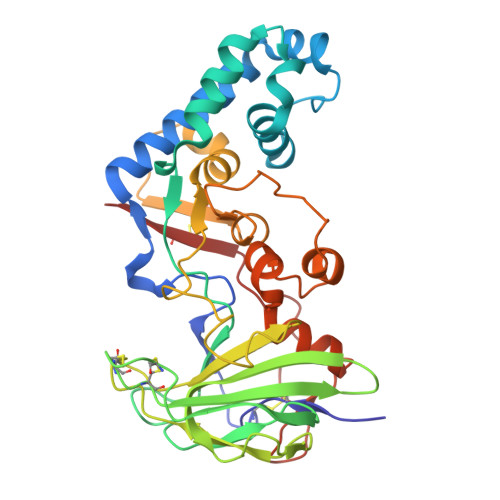
Crystal structure, enzymatic and thermodynamic properties of the Thermus thermophilus phage Tt72 lytic endopeptidase with unique structural signatures of thermal adaptation.
Dorawa, S., Biniek-Antosiak, K., Bejger, M., Kaczorowska, A.K., Ciuchcinski, K., Godlewska, A., Plotka, M., Hreggvidsson, G.O., Dziewit, L., Kaczorowski, T., Rypniewski, W.(2025) J Struct Biol 217: 108230-108230
- PubMed: 40580998
- DOI: https://doi.org/10.1016/j.jsb.2025.108230
- Primary Citation of Related Structures:
8S3M, 8S3U, 8S3W - PubMed Abstract:
We presents the discovery and molecular characterization of a novel lytic enzyme from the extremophilic Thermus thermophilus MAT72 phage vB_Tt72. The protein of 346-aa (MW = 39,705) functions as phage vB_Tt72 endolysin and shows low sequence identity (<37 %) to members of M23 family of peptidoglycan hydrolases, except for two uncharacterized endopeptidases of T. thermophilus phages: φYS40 (87 %) and φTMA (88 %). The enzyme exhibits lytic activity mainly against bacteria of the genus Thermus and, to a lesser extent, against other Gram-negative and Gram-positive bacteria. The protein is monomeric in solution and is highly thermostable (T m = 98.3 °C). It retains ∼ 50 % of its lytic activity after 90 min of incubation at 99 °C. Crystallographic analysis, at 2.2 Å resolution, revealed a fold characteristic of M23 metallopeptidases, accounting for 40 % of the structure. The remaining parts of the molecule are folded in a manner that was previously undescribed. The M23 fold contains a Zn 2+ ion coordinated by a conserved His-Asp-His triad, and two conserved His residues essential for catalysis. The active site is occupied by a phosphate or a sulfate anion, while the substrate-binding groove contains a ligand, which is a fragment of E. coli peptidoglycan. The common sequence-based criteria failed to identify the protein as (hyper)thermophilic. It is likely that the protein's thermal stability is owed to peculiar features of its three-dimensional structure. Instead of trimmed surface loops, observed in many thermostable proteins, the catalytic domain contains two long loops that interlace and form an α-helical bundle with its own hydrophobic core.
- Laboratory of Extremophiles Biology, Department of Microbiology, Faculty of Biology, University of Gdansk, 80-308 Gdańsk, Poland.
Organizational Affiliation: