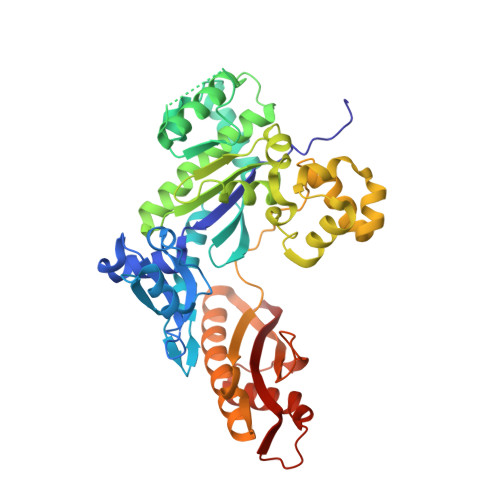
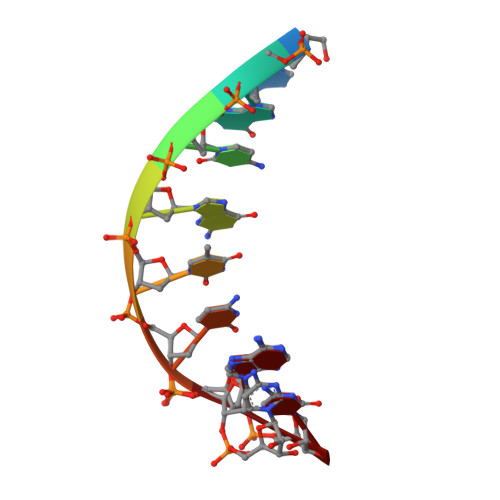
Sugar ring alignment and dynamics underline cytarabine and gemcitabine inhibition on Pol eta catalyzed DNA synthesis.
Chang, C., Zhou, G., Lee Luo, C., Eleraky, S., Moradi, M., Gao, Y.(2024) J Biological Chem 300: 107361-107361
- PubMed: 38735473
- DOI: https://doi.org/10.1016/j.jbc.2024.107361
- Primary Citation of Related Structures:
8V7A, 8V7B, 8V7C, 8V7D, 8V7E, 8V7F, 8V7G, 8V7H, 8V7I, 8V7J, 8V7K - PubMed Abstract:
Nucleoside analogue drugs are pervasively used as antiviral and chemotherapy agents. Cytarabine and gemcitabine are anti-cancer nucleoside analogue drugs that contain C2' modifications on the sugar ring. Despite carrying all the required functional groups for DNA synthesis, these two compounds inhibit DNA extension once incorporated into DNA. It remains unclear how the C2' modifications on cytarabine and gemcitabine affect the polymerase active site during substrate binding and DNA extension. Using steady-state kinetics, static and time-resolved X-ray crystallography with DNA polymerase η (Pol η) as a model system, we showed that the sugar ring C2' chemical groups on cytarabine and gemcitabine snugly fit within the Pol η active site without occluding the steric gate. During DNA extension, Pol η can extend past gemcitabine but with much lower efficiency past cytarabine. The Pol η crystal structures show that the -OH modification in the β direction on cytarabine locks the sugar ring in an unfavorable C2'-endo geometry for product formation. On the other hand, the addition of fluorine atoms on gemcitabine alters the proper conformational transition of the sugar ring for DNA synthesis. Our study illustrates mechanistic insights into chemotherapeutic drug inhibition and resistance and guides future optimization of nucleoside analogue drugs.
- Department of Biosciences, Rice University, Houston, Texas, USA.
Organizational Affiliation: