Ellagic acid metabolism as a source of dietary MR1 ligands
Wang, C.J.H., Tang, J.S., Stephens, R., Faulkner, S., Gell, K., Li, Y., Cait, A., Rossjohn, J., Le Nours, J., Gasser, O.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Major histocompatibility complex class I-related gene protein | 271 | Homo sapiens | Mutation(s): 0 Gene Names: MR1 | 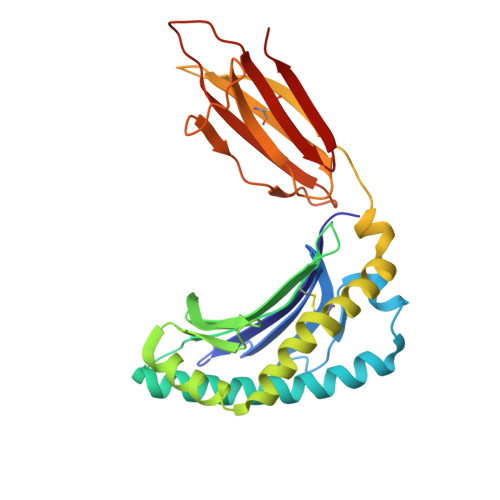 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q95460 (Homo sapiens) Explore Q95460 Go to UniProtKB: Q95460 | |||||
PHAROS: Q95460 GTEx: ENSG00000153029 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q95460 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | 100 | Homo sapiens | Mutation(s): 0 Gene Names: B2M, CDABP0092, HDCMA22P | 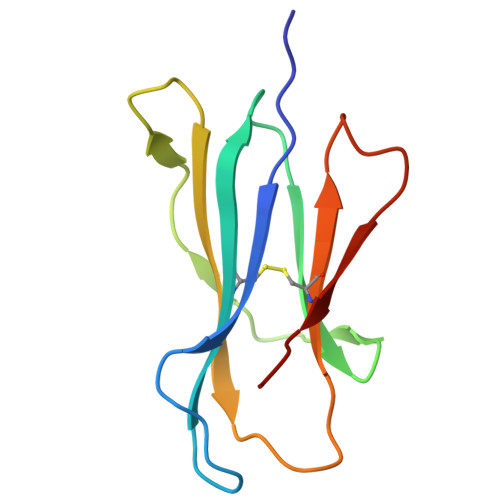 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61769 (Homo sapiens) Explore P61769 Go to UniProtKB: P61769 | |||||
PHAROS: P61769 GTEx: ENSG00000166710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61769 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TRA@ protein | 204 | Homo sapiens | Mutation(s): 0 Gene Names: TRA@ | 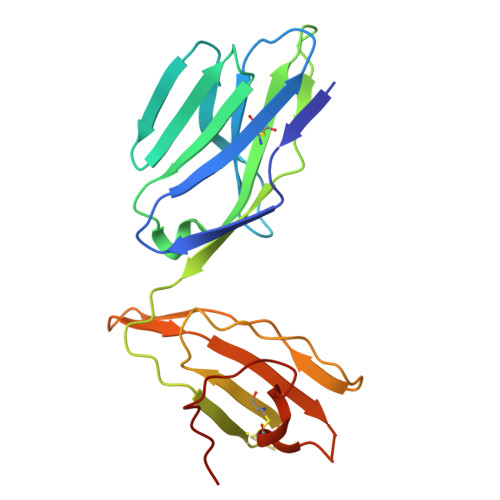 | |
UniProt | |||||
Find proteins for Q6P4G7 (Homo sapiens) Explore Q6P4G7 Go to UniProtKB: Q6P4G7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6P4G7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MAIT T-cell receptor beta chain | 246 | Homo sapiens | Mutation(s): 0 | 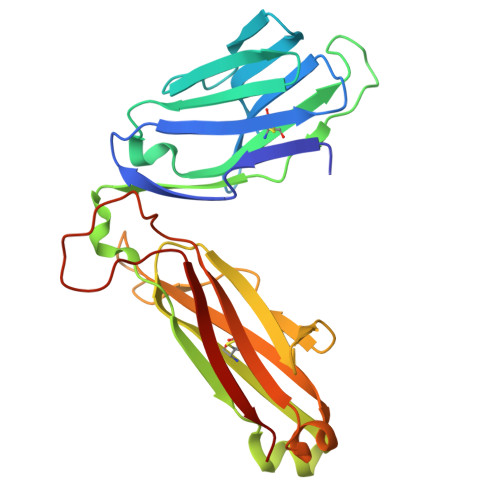 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| REF (Subject of Investigation/LOI) Query on REF | I [auth A], M [auth C] | 2,3,7,8-tetrahydroxychromeno[5,4,3-cde]chromene-5,10-dione C14 H6 O8 AFSDNFLWKVMVRB-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | Q [auth E], R [auth F], S [auth F] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| ACY Query on ACY | L [auth A], O [auth C], P [auth D] | ACETIC ACID C2 H4 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-N |  | ||
| CL Query on CL | N [auth C] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | J [auth A], K [auth A], T [auth H] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 215.81 | α = 90 |
| b = 70.352 | β = 104.09 |
| c = 143.434 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Health and Medical Research Council (NHMRC, Australia) | Australia | 2008981 |