Discovery of a novel alkyne-based scaffold inhibitor for human DHODH.
Purificacao, A.D., Vaidergorn, M.M., Silva-Mendonca, S., Silva, W.J.L., Leite, P.I.P., Santos, T., Andrade, C.H., Nonato, M.C., Emery, F.S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dihydroorotate dehydrogenase (quinone), mitochondrial | 368 | Homo sapiens | Mutation(s): 0 Gene Names: DHODH EC: 1.3.5.2 | 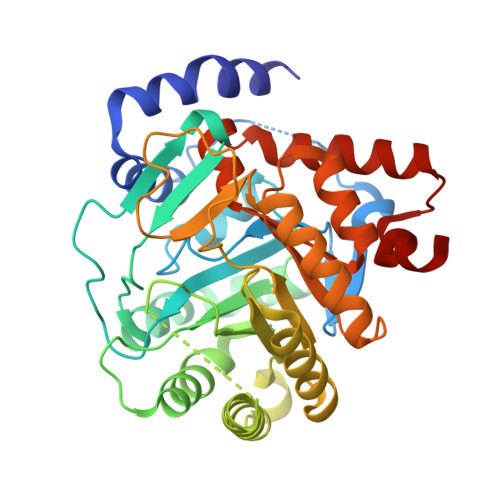 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q02127 (Homo sapiens) Explore Q02127 Go to UniProtKB: Q02127 | |||||
PHAROS: Q02127 GTEx: ENSG00000102967 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q02127 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FMN (Subject of Investigation/LOI) Query on FMN | I [auth A] | FLAVIN MONONUCLEOTIDE C17 H21 N4 O9 P FVTCRASFADXXNN-SCRDCRAPSA-N |  | ||
| A1AVV (Subject of Investigation/LOI) Query on A1AVV | C [auth A] | N-{4-[(3-aminophenyl)ethynyl]phenyl}acetamide C16 H14 N2 O GDYCMPAYKROXHM-UHFFFAOYSA-N |  | ||
| ORO (Subject of Investigation/LOI) Query on ORO | B [auth A] | OROTIC ACID C5 H4 N2 O4 PXQPEWDEAKTCGB-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | D [auth A], H [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | E [auth A], F [auth A], J [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CL Query on CL | G [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 91.042 | α = 90 |
| b = 91.042 | β = 90 |
| c = 122.616 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| pointless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Brazilian National Council for Scientific and Technological Development (CNPq) | Brazil | 441038/2020-4 |
| Sao Paulo Research Foundation (FAPESP) | Brazil | 2020/06190-0 |
| Sao Paulo Research Foundation (FAPESP) | Brazil | 2021/13237-5 |