Covalent Targeting of Histidine Residues with Aryl Fluorosulfates: Application to Mcl-1 BH3 Mimetics.
Alboreggia, G., Udompholkul, P., Atienza, E.L., Muzzarelli, K., Assar, Z., Pellecchia, M.(2024) J Med Chem 67: 20214-20223
- PubMed: 39532346
- DOI: https://doi.org/10.1021/acs.jmedchem.4c01541
- Primary Citation of Related Structures:
9CKN - PubMed Abstract:
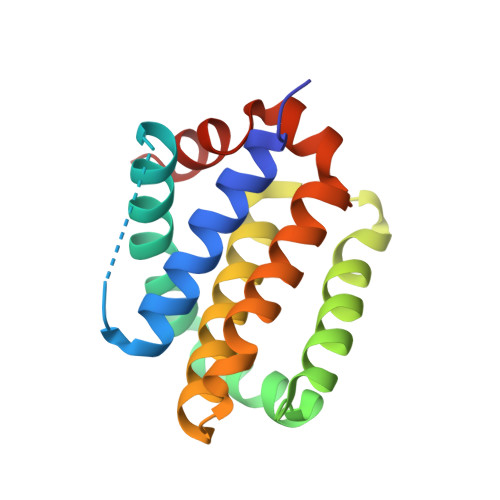
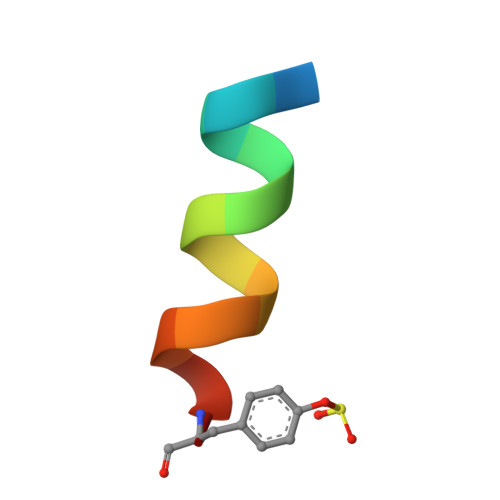
Covalent drugs provide pharmacodynamic and pharmacokinetic advantages over reversible agents. However, covalent strategies have been developed mostly to target cysteine (Cys) residues, which are rarely found in binding sites. Among other nucleophilic residues that could be in principle used for the design of covalent drugs, histidine (His) has not been given proper attention despite being in principle an attractive residue to pursue but underexplored. Aryl fluorosulfates, a mild electrophile that is very stable in biological media, have been recently identified as possible electrophiles to react with the side chains of Lys; however, limited studies are available on aryl fluorosulfates' ability to target His residues. We demonstrate that proper incorporation of an aryl fluorosulfate juxtaposing the electrophile with a His residue can be used to afford rapid optimizations of His-covalent agents. As an application, we report on His-covalent BH3 mimetics targeting His224 of Mcl-1.
Organizational Affiliation:
Division of Biomedical Sciences, School of Medicine, University of California Riverside, 900 University Avenue, Riverside, California 92521, United States.