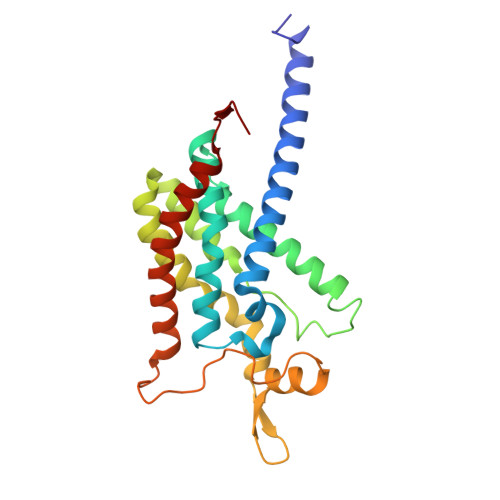
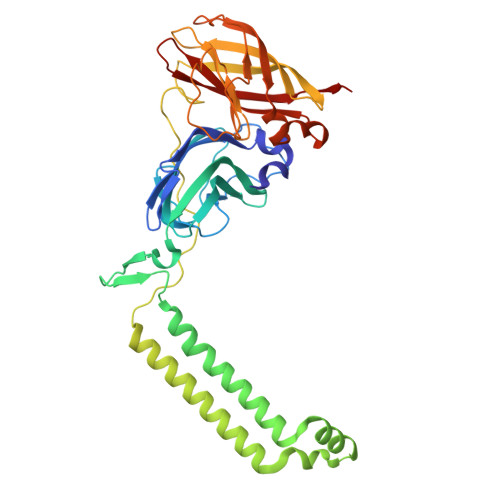
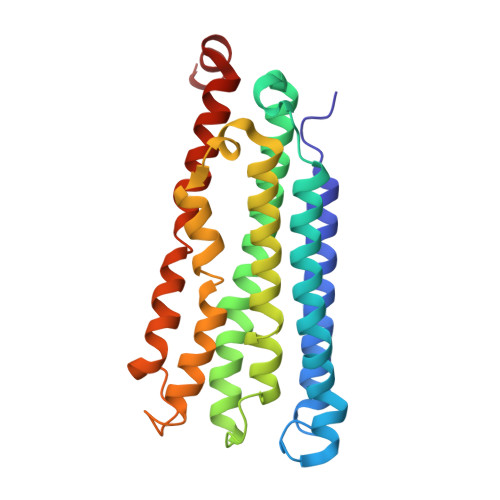
Structures of methane and ammonia monooxygenases in native membranes.
Tucci, F.J., Rosenzweig, A.C.(2025) Proc Natl Acad Sci U S A 122: e2417993121-e2417993121
- PubMed: 39739801
- DOI: https://doi.org/10.1073/pnas.2417993121
- Primary Citation of Related Structures:
9CL1, 9CL2, 9CL3, 9CL4, 9CL5, 9CL6 - PubMed Abstract:
Methane- and ammonia-oxidizing bacteria play key roles in the global carbon and nitrogen cycles, respectively. These bacteria use homologous copper membrane monooxygenases to accomplish the defining chemical transformations of their metabolisms: the oxidations of methane to methanol by particulate methane monooxygenase (pMMO) and ammonia to hydroxylamine by ammonia monooxygenase (AMO), enzymes of prime interest for applications in mitigating climate change. However, investigations of these enzymes have been hindered by the need for disruptive detergent solubilization prior to structure determination, confounding studies of pMMO and precluding studies of AMO. Here, we overcome these challenges by using cryoEM to visualize pMMO and AMO directly in their native membrane arrays at 2.4 to 2.8 Å resolution. These structures reveal details of the copper centers, numerous bound lipids, and previously unobserved components, including identifiable and distinct supernumerary helices interacting with pMMO and AMO, suggesting a widespread role for these helices in copper membrane monooxygenases. Comparisons between these structures, their metallocofactors, and their unexpected protein-protein interactions highlight features that may govern activity or the formation of higher-order arrays in native membranes. The ability to obtain molecular insights within the native membrane will enable further understanding of these environmentally important enzymes.
- Departments of Molecular Biosciences and of Chemistry, Northwestern University, Evanston, IL 60208.
Organizational Affiliation: