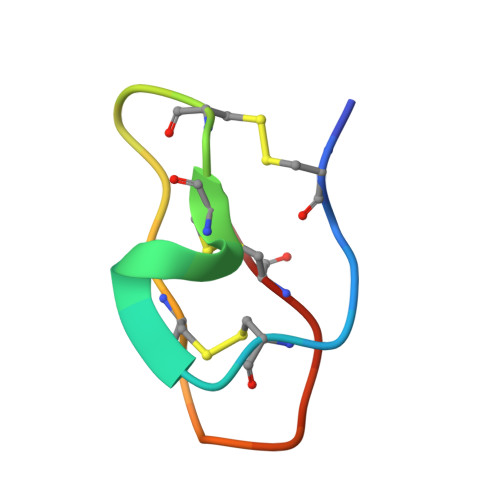
Structure and functional studies of Avt1, a novel peptide from the sea anemone Aulactinia veratra.
Albar, R.A., Smith, H.L., Sanches, K., Wai, D.C.C., Naseem, M.U., Szanto, T.G., Panyi, G., Prentis, P.J., Norton, R.S.(2024) Biochim Biophys Acta Proteins Proteom 1873: 141050-141050
- PubMed: 39357665
- DOI: https://doi.org/10.1016/j.bbapap.2024.141050
- Primary Citation of Related Structures:
9CRB - PubMed Abstract:
Sea anemones are a rich source of peptide toxins spanning a diverse range of biological activities, typically targeting proteins such as ion channels, receptors and transporters. These peptide toxins and their analogues are usually highly stable and selective for their molecular targets, rendering them of interest as molecular tools, insecticides and therapeutics. Recent transcriptomic and proteomic analyses of the sea anemone Aulactinia veratra identified a novel 28-residue peptide, designated Avt1. Avt1 was produced using solid-phase peptide synthesis, followed by oxidative folding and purification of the folded peptide using reversed-phase high-performance liquid chromatography. The liquid chromatography-mass spectrometry profile of synthetic Avt1 showed a pure peak with molecular mass 6 Da less than that of the reduced form of the peptide, indicating the successful formation of three disulfide bonds. The solution structure determined by NMR revealed that Avt1 adopts an inhibitor cystine knot (ICK) fold, in which a ring is formed by two disulfide bonds with a third disulfide penetrating the ring to create the pseudo-knot. This structure provides ICK peptides with high structural, thermal and proteolytic stability. Consistent with its ICK structure, Avt1 was resistant to proteolysis by trypsin, chymotrypsin and pepsin, although it was not a trypsin inhibitor. Avt1 at 100 nM showed no activity in patch-clamp electrophysiological assays against several mammalian voltage-gated ion channels, but has structural features similar to toxins targeting insect sodium ion channels. Although sequence homologues of Avt1 are found in a number of sea anemones, this is the first representative of this family to be characterised structurally and functionally.
- Medicinal Chemistry, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, VIC 3052, Australia.
Organizational Affiliation: