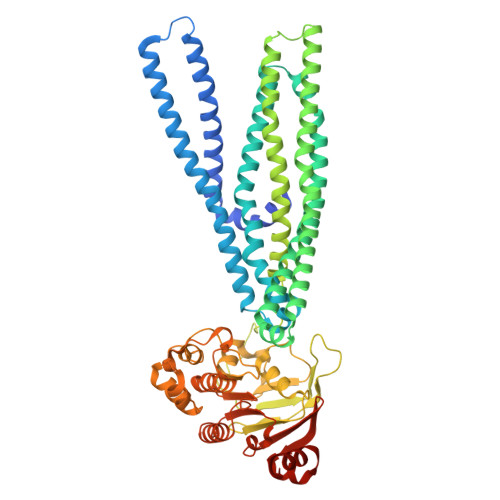
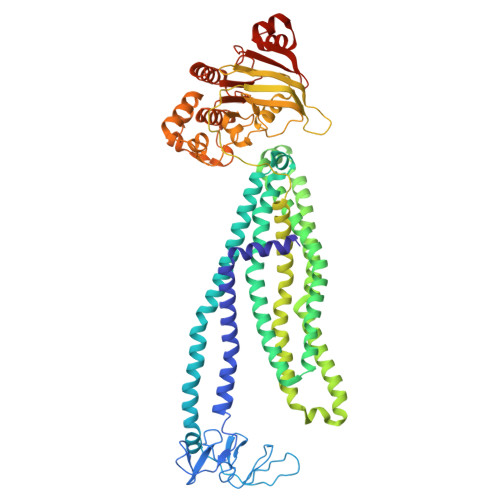
Lipid-mediated mechanism of drug extrusion by a heterodimeric ABC exporter.
Tang, Q., Sinclair, M., Bisignano, P., Zhang, Y., Tajkhorshid, E., Mchaourab, H.S.(2025) bioRxiv
- PubMed: 40060395
- DOI: https://doi.org/10.1101/2025.02.26.640354
- Primary Citation of Related Structures:
9CUP, 9CUR, 9CUS - PubMed Abstract:
Multidrug transport by ATP binding cassette (ABC) exporters entails a mechanism to modulate drug affinity across the transport cycle. Here, we combine cryo-EM and molecular dynamics (MD) simulations to illuminate how lipid competition modulates substrate affinity to drive its translocation by ABC exporters. We determined cryo-EM structures of the ABC transporter BmrCD in drug-loaded inward-facing (IF) and outward-facing (OF) conformations in lipid nanodiscs to reveal the structural basis of alternating access, details of drug-transporter interactions, and the scale of drug movement between the two conformations. Remarkably, the structures uncovered lipid molecules bound in or near the transporter vestibule along with the drugs. MD trajectories from the IF structure show that these lipids stimulate drug disorder and translocation towards the vestibule apex. Similarly, bound lipids enter the OF vestibule and weaken drug-transporter interactions facilitating drug release. Our results complete a near-atomic model of BmrCD's conformational cycle and advance a general mechanism of lipid-driven drug transport by ABC exporters.
- Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN 37232,USA.
Organizational Affiliation: