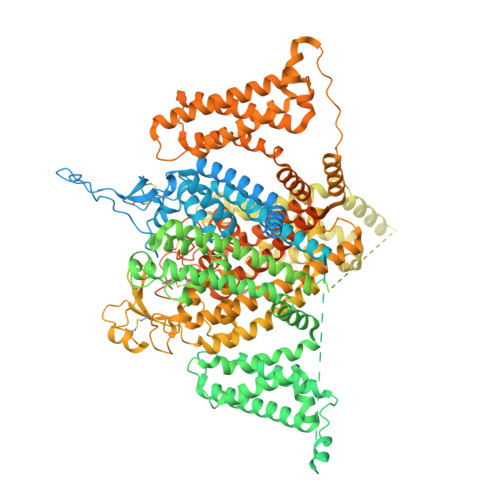
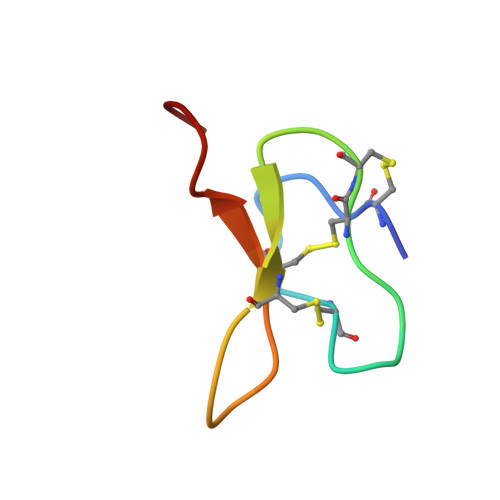
Structural basis of inhibition of human Na V 1.8 by the tarantula venom peptide Protoxin-I.
Neumann, B., McCarthy, S., Gonen, S.(2025) Nat Commun 16: 1459-1459
- PubMed: 39920100
- DOI: https://doi.org/10.1038/s41467-024-55764-z
- Primary Citation of Related Structures:
9DBK, 9DBL, 9DBM, 9DBN - PubMed Abstract:
Voltage-gated sodium channels (Na V s) selectively permit diffusion of sodium ions across the cell membrane and, in excitable cells, are responsible for propagating action potentials. One of the nine human Na V isoforms, Na V 1.8, is a promising target for analgesics, and selective inhibitors are of interest as therapeutics. One such inhibitor, the gating-modifier peptide Protoxin-I derived from tarantula venom, blocks channel opening by shifting the activation voltage threshold to more depolarized potentials, but the structural basis for this inhibition has not previously been determined. Using monolayer graphene grids, we report the cryogenic electron microscopy structures of full-length human apo-Na V 1.8 and the Protoxin-I-bound complex at 3.1 Å and 2.8 Å resolution, respectively. The apo structure shows an unexpected movement of the Domain I S4-S5 helix, and VSD I was unresolvable. We find that Protoxin-I binds to and displaces the VSD II S3-S4 linker, hindering translocation of the S4 II helix during activation.
- Department of Molecular Biology and Biochemistry, University of California Irvine, Irvine, CA, USA.
Organizational Affiliation: