Chemical Evolution of Aplithianine Class of Serine/Threonine Kinase Inhibitors.
Du, L., Wilson, B.A.P., Moore, W.J., Dalilian, M., Shenoy, S.R., Li, N., Martinez Fiesco, J.A., Hwang, J.Y., Smith, E.A., Wamiru, A., Goncharova, E.I., Alvarez de la Cruz, A., Pagadala, K., Piswa, H.K., Patteti, V., Jampana, V.P., Manepalli, P., Nimmala, R., Marri, N.R., Gunuguntla, M., Reddy, J.J., Schneekloth Jr., J.S., Zhang, P., O'Keefe, B.R.(2025) J Med Chem 68: 12756-12785
- PubMed: 40476486
- DOI: https://doi.org/10.1021/acs.jmedchem.5c00649
- Primary Citation of Related Structures:
9DC6, 9DCD - PubMed Abstract:
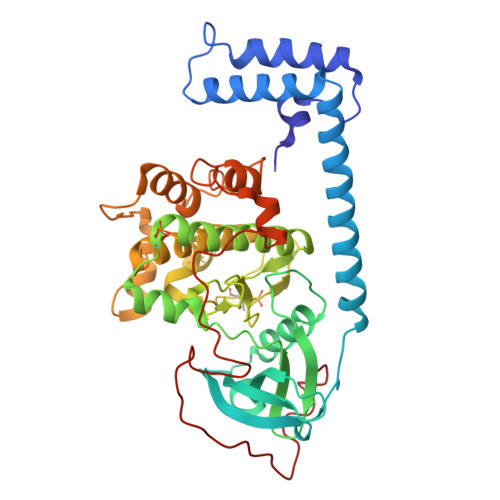
Chimeric kinase J-PKAcα represents a potential therapeutic target for fibrolamellar hepatocellular carcinoma (FLHCC). Structure-based design and screening were applied to improve the potency of the marine-derived kinase inhibitor aplithianine A targeting J-PKAcα. Three classes of aplithianines (I, II, and III) including >150 analogs were synthesized, significantly improving biochemical IC 50 values to the low nanomolar range. X-ray diffraction experiments confirmed that the class II aplithianines adopted a novel binding mode to J-PKAcα by interacting with the DFG residue Asp239. The kinase selectivity profiles were assessed by kinome profiling. In vitro profiles of selected class II analogs were evaluated to determine compound solubility, protein binding, permeability, metabolism, and hERG binding. Selected aplithianine analogs inhibited intracellular phosphorylation of the peptide substrate CREB following stimulation of the J-PKAcα fusion kinase in NIH/3T3 cells and exhibited antiproliferative/cytotoxic activities against select cancer cell lines from the NCI-60 cell panel at nanomolar concentrations.
- Molecular Targets Program, Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702, United States.
Organizational Affiliation: