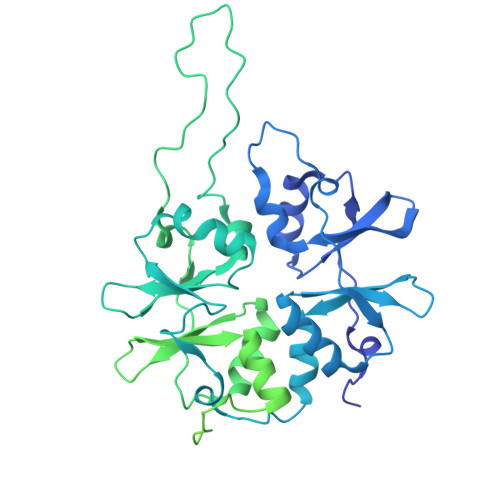
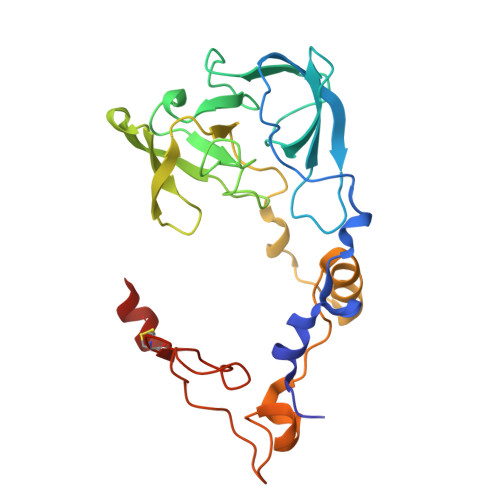
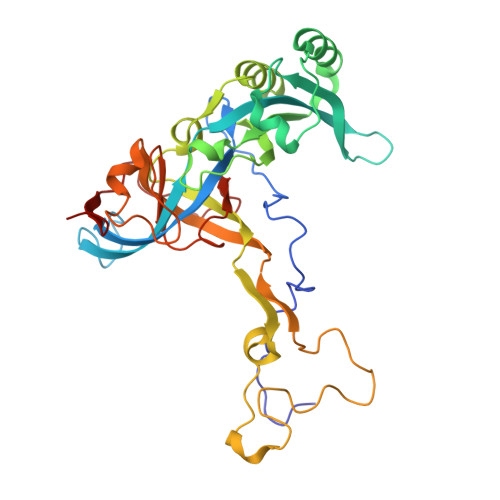
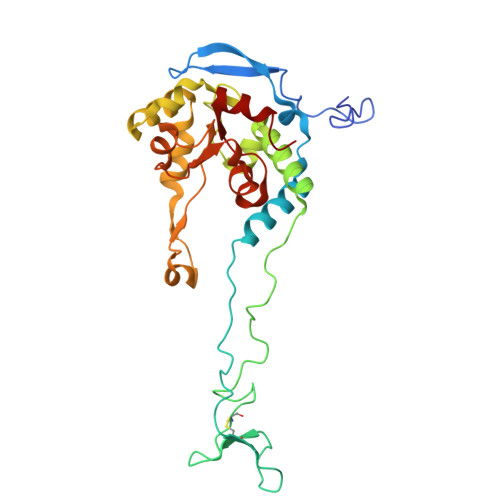
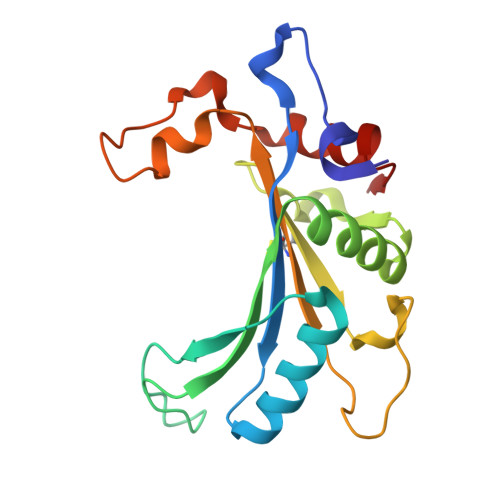
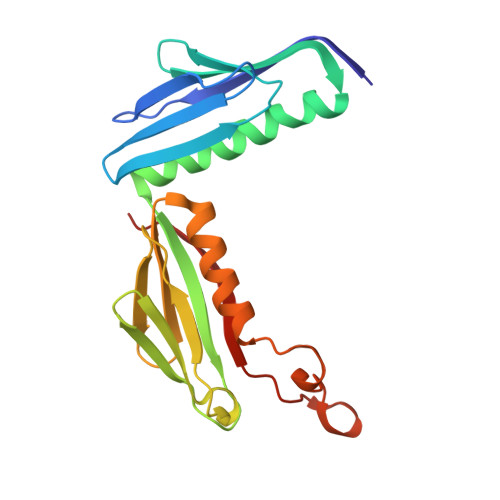
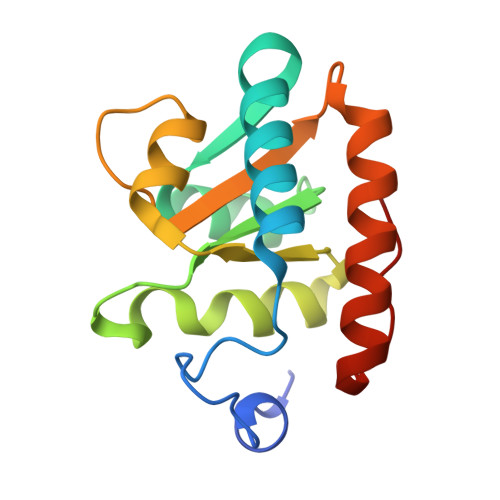
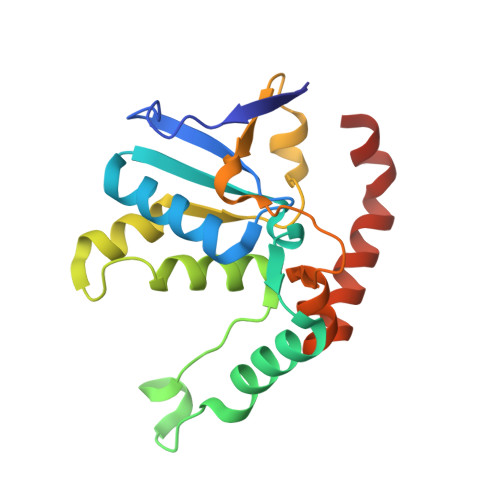
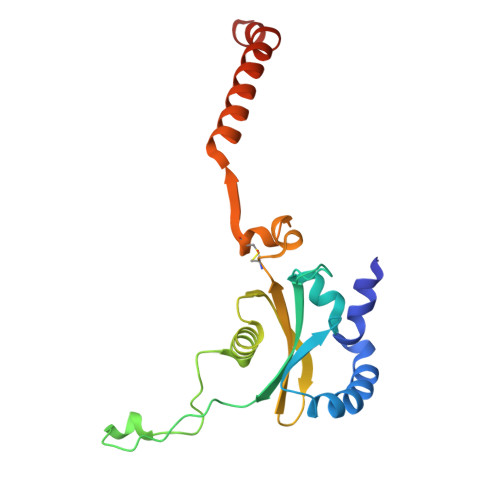
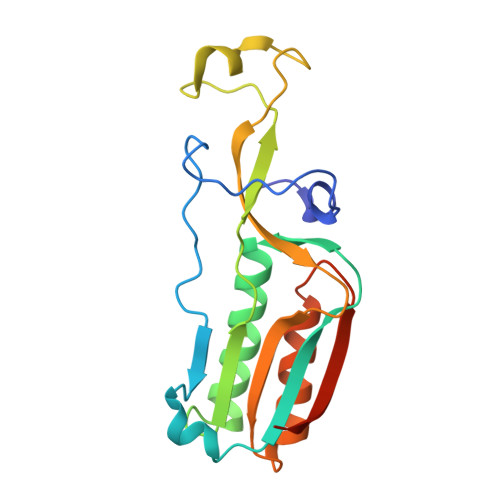
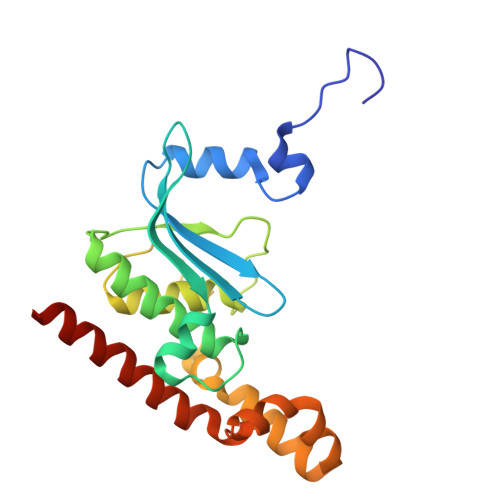
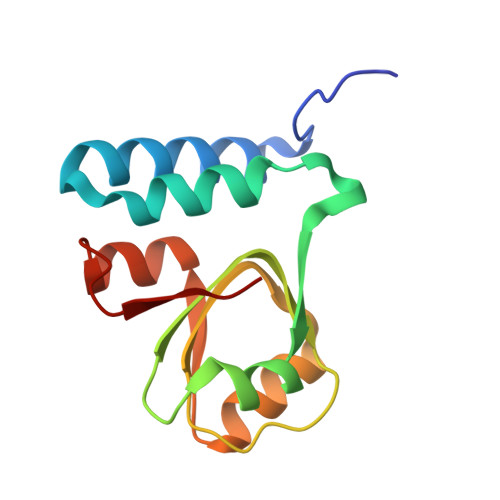
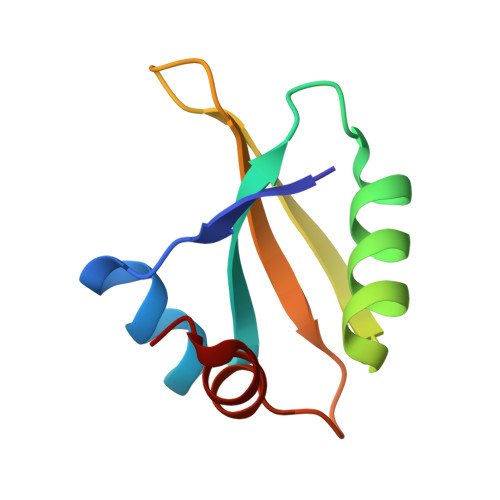
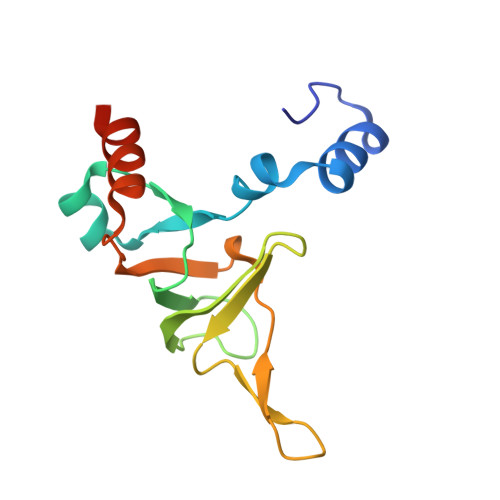
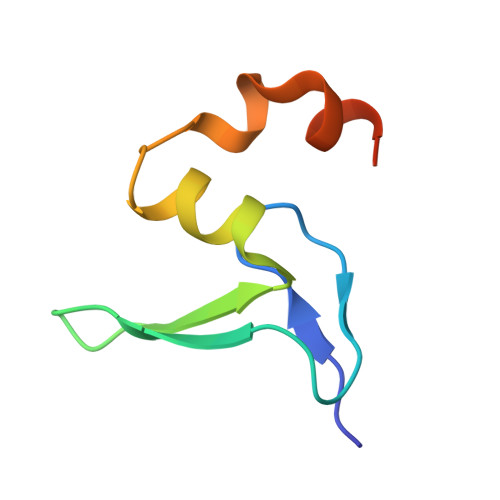
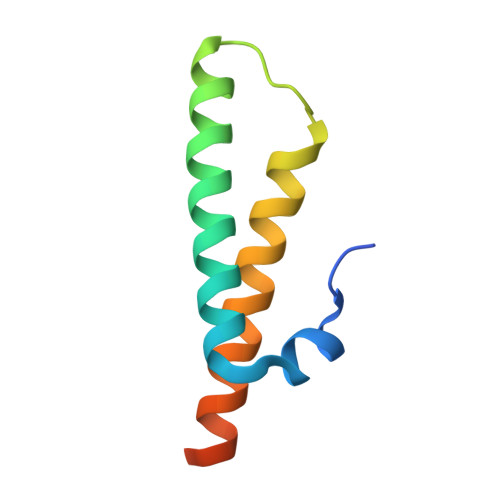
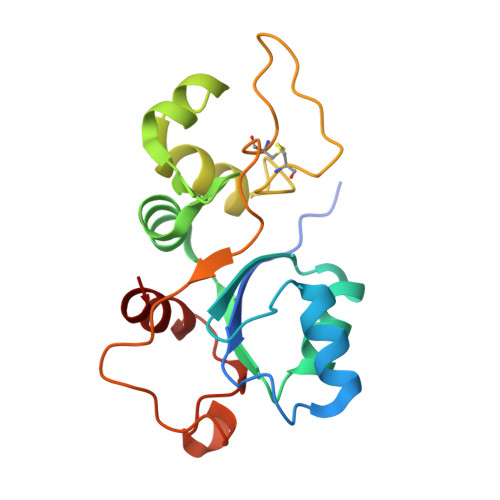
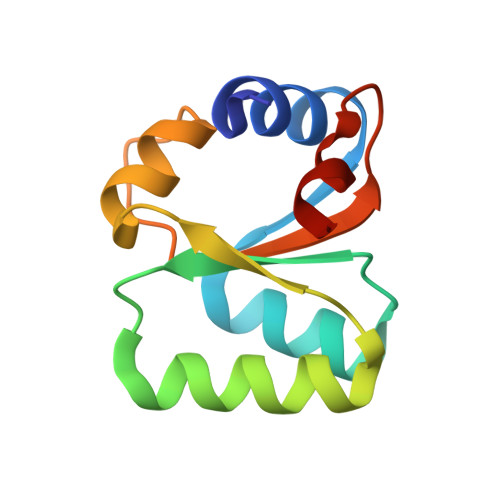
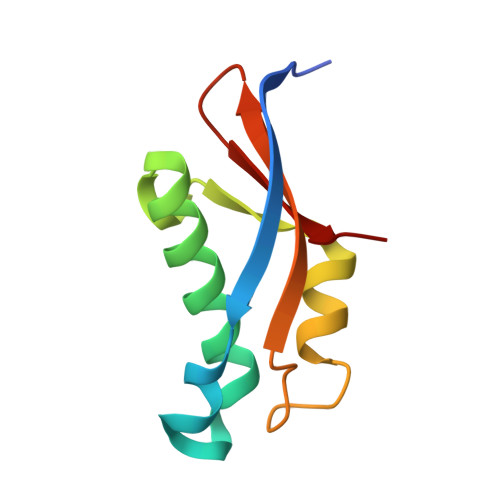
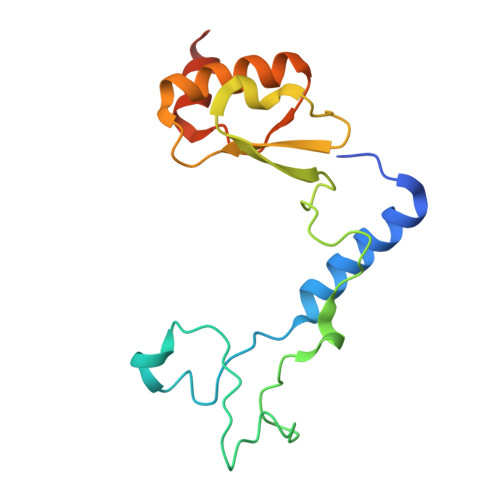
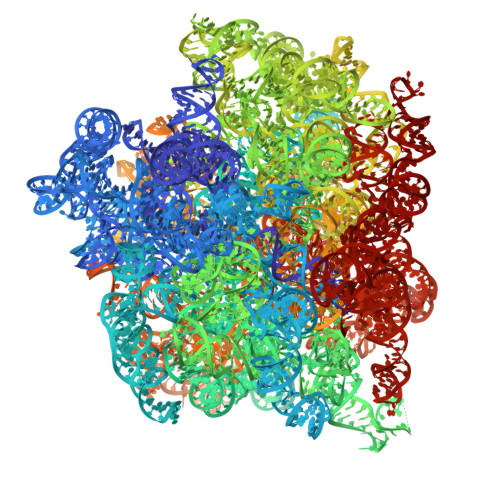
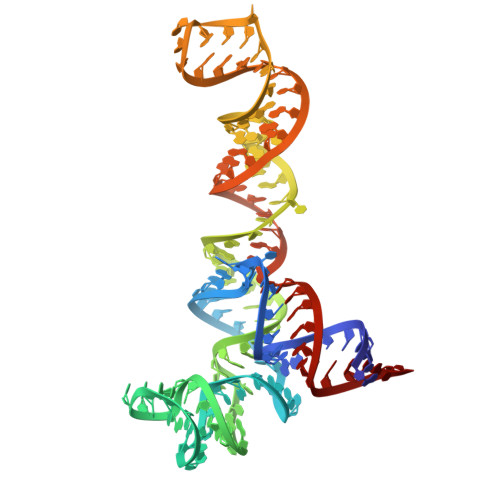
Structure of an archaeal ribosome reveals a divergent active site and hibernation factor.
Nissley, A.J., Shulgina, Y., Kivimae, R.W., Downing, B.E., Penev, P.I., Banfield, J.F., Nayak, D.D., Cate, J.H.D.(2025) Nat Microbiol 10: 1940-1953
- PubMed: 40676158
- DOI: https://doi.org/10.1038/s41564-025-02065-w
- Primary Citation of Related Structures:
9E6Q, 9E71, 9E7F - PubMed Abstract:
Ribosomes translate mRNA into protein. Despite divergence in ribosome structure over the course of evolution, the catalytic site, known as the peptidyl transferase centre (PTC), is thought to be nearly universally conserved. Here we identify clades of archaea that have highly divergent ribosomal RNA sequences in the PTC. To understand how these PTC sequences fold, we determined cryo-EM structures of the 70S and 50S ribosomes to 2.4 Å and 2 Å, respectively, from the hyperthermophilic archaeon Pyrobaculum calidifontis. PTC sequence variation leads to the rearrangement of key base triples, and differences between archaeal and bacterial ribosomal proteins enable sequence variation in archaeal PTCs. Finally, we identify an archaeal ribosome hibernation factor, Dri, that differs from known bacterial and eukaryotic hibernation factors and is found in multiple archaeal phyla. Overall, this work identifies factors that regulate ribosome function in archaea and reveals a larger diversity of the most ancient sequences in the ribosome.
- Department of Chemistry, University of California, Berkeley, Berkeley, CA, USA.
Organizational Affiliation: