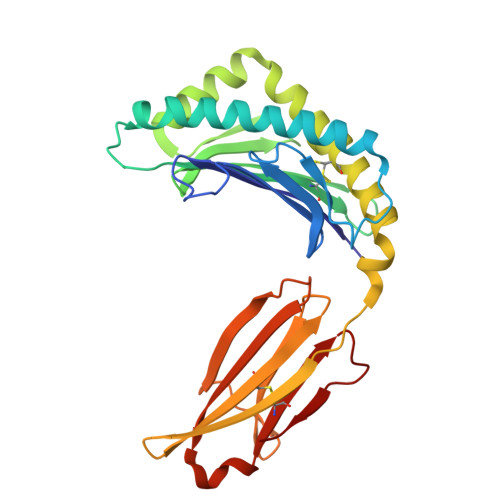
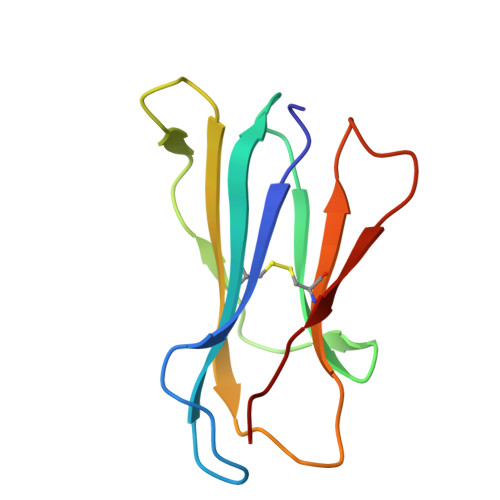
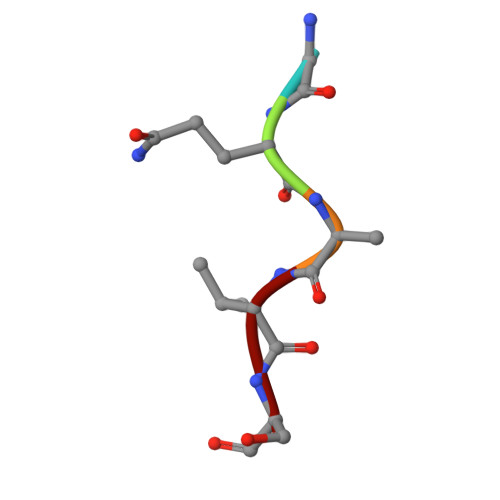
Molecular basis for presentation of N-myristoylated peptides by the chicken YF1∗7.1 molecule.
Khandokar, Y., Cheng, T.Y., Wang, C.J.H., Cao, T.P., Nagampalli, R.S.K., Sivaraman, K.K., Van Rhijn, I., Rossjohn, J., Moody, D.B., Nours, J.L.(2025) J Biological Chem 301: 110253-110253
- PubMed: 40412526
- DOI: https://doi.org/10.1016/j.jbc.2025.110253
- Primary Citation of Related Structures:
9EB2, 9EB3, 9EB4, 9EB5, 9EB6 - PubMed Abstract:
Major histocompatibility complex I (MHC-I) and MHC-I-like molecules play a central role in mediating immunity. Through their conservation across all taxa of jawed vertebrates, the MHC-I-like proteins have adapted to present non-peptidic antigens to distinct T cell populations. While our understanding of the structure-function relationship of MHC-I and MHC-I-like molecules in humans and mice is well established, the nature of the antigens presented by MHC-I- like molecules in "non-model" species remains unclear. Here, using a mammalian recombinant expression system combined with mass spectrometry approaches, we identified N-myristoylated peptides as endogenous ligands for the chicken MHC-I-like protein YF1∗7.1. Given the importance of N-myristoylation in viral pathogenesis, we determined the crystal structure of YF1∗7.1 in complex with two N-myristoylated peptides derived from Marek's disease virus (MDV), demonstrating the molecular basis that underpins the presentation of N-myristoylated peptides from MDV, a highly contagious and fatal viral neoplastic disease in chickens. Thus, the identified ligands are distinct from unmodified peptides found in classical MHC-I and -II as well as diverse amphipathic lipids captured by CD1 proteins. Collectively, our study lays the foundation for further molecular and functional characterization of YF1∗7.1 and more broadly of the role of the MHC-I encoded by the MHC-Y gene cluster in protection against highly contagious viral neoplastic diseases in chickens.
- Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: