Structure and mechanism of haem-dependent nitrogen-nitrogen bond formation in piperazate synthase
Higgins, M.A., Shi, X., Soler, J., Harland, J.B., Parkkila, T., Lehnert, N., Garcia-Borras, M., Du, Y.L., Ryan, K.S.(2025) Nat Catal
Experimental Data Snapshot
Starting Model: experimental
View more details
(2025) Nat Catal
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Piperazate synthase | A [auth B], B [auth A] | 226 | Streptomyces griseus subsp. griseus | Mutation(s): 0 | 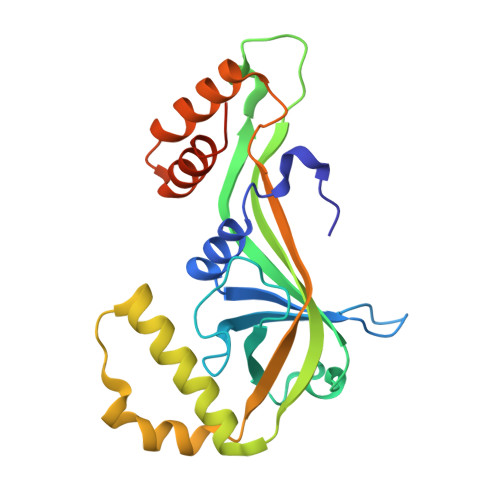 |
UniProt | |||||
Find proteins for A0A7K2YFL1 (Streptomyces sp. SID3343) Explore A0A7K2YFL1 Go to UniProtKB: A0A7K2YFL1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7K2YFL1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM (Subject of Investigation/LOI) Query on HEM | C [auth B], E [auth A] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| ONH (Subject of Investigation/LOI) Query on ONH | D [auth B], H [auth A] | N~5~-hydroxy-L-ornithine C5 H12 N2 O3 OZMJDTPATROLQC-BYPYZUCNSA-N |  | ||
| GOL Query on GOL | F [auth A], G [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 67.002 | α = 90 |
| b = 67.002 | β = 90 |
| c = 479.001 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Natural Sciences and Engineering Research Council (NSERC, Canada) | Canada | RPGIN-2021-02626 |