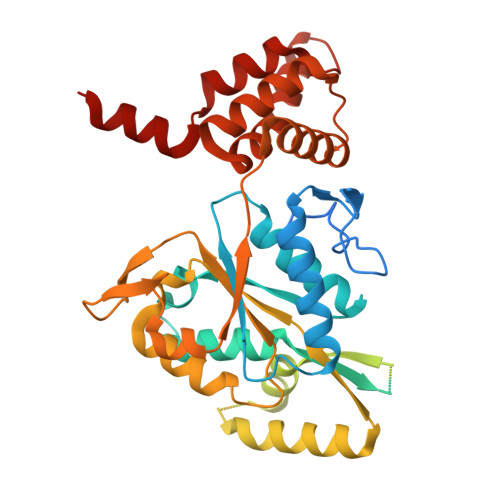
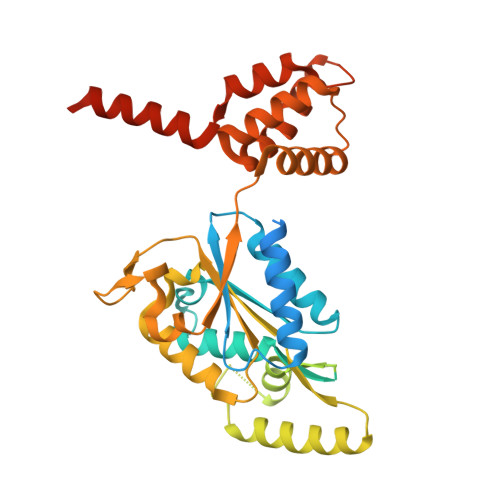
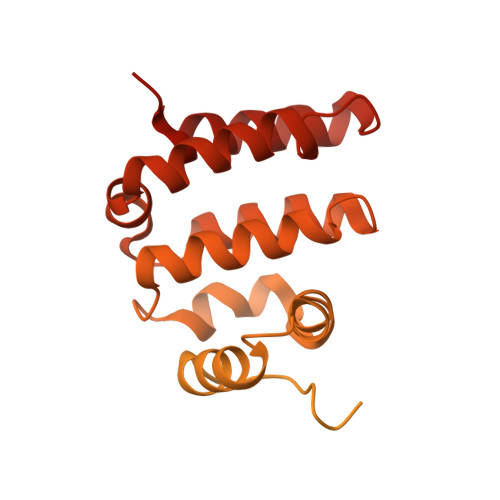
The structure of the R2T complex reveals a different architecture from the related HSP90 cochaperone R2TP.
Palacios-Abella, A., Lopez-Perrote, A., Boskovic, J., Fonseca, S., Urbez, C., Rubio, V., Llorca, O., Alabadi, D.(2025) Structure 33: 740
- PubMed: 40015274
- DOI: https://doi.org/10.1016/j.str.2025.01.023
- Primary Citation of Related Structures:
9EQ2 - PubMed Abstract:
The R2TP complex is a specialized HSP90 cochaperone essential for the maturation of macromolecular complexes such as RNAPII and TORC1. R2TP is formed by a hetero-hexameric ring of AAA-ATPases RuvBL1 and RuvBL2, which interact with RPAP3 and PIH1D1. Several R2TP-like complexes have been described, but these are less well characterized. Here, we identified, characterized and determined the cryo-electron microscopy (cryo-EM) structure of R2T from Arabidopsis thaliana, which lacks PIH1D1 and is probably the only form of the complex in seed plants. In contrast to R2TP, R2T is organized as two rings of AtRuvBL1-AtRuvBL2a interacting back-to-back, with one AtRPAP3 anchored per ring. AtRPAP3 has no effect on the ATPase activity of AtRuvBL1-AtRuvBL2a and binds with a different stoichiometry than in human R2TP. We show that the interaction of AtRPAP3 with AtRuvBL2a and AtHSP90 occurs via a conserved mechanism. However, the distinct architectures of R2T and R2TP suggest differences in their functions and mechanisms.
Organizational Affiliation:
Instituto de Biología Molecular y Celular de Plantas (CSIC-UPV), Ingeniero Fausto Elio s/n, 46022 Valencia, Spain.