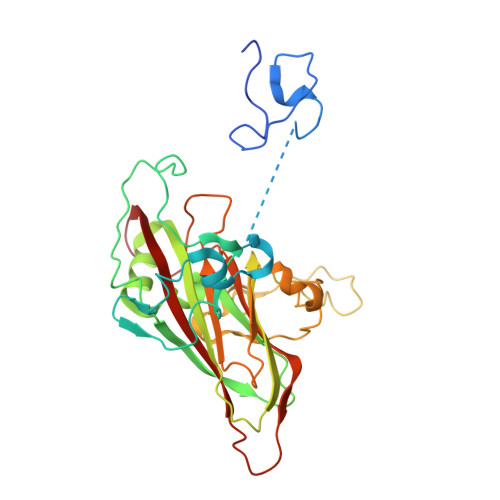
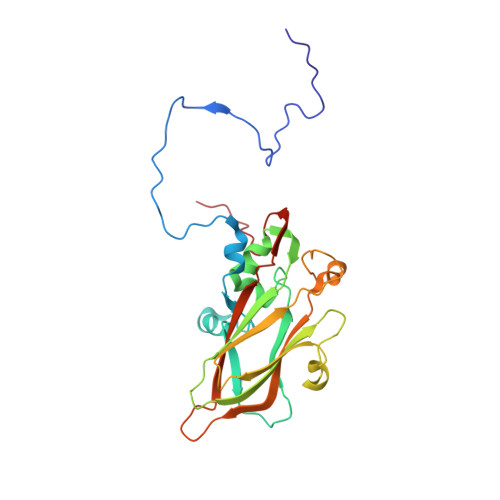
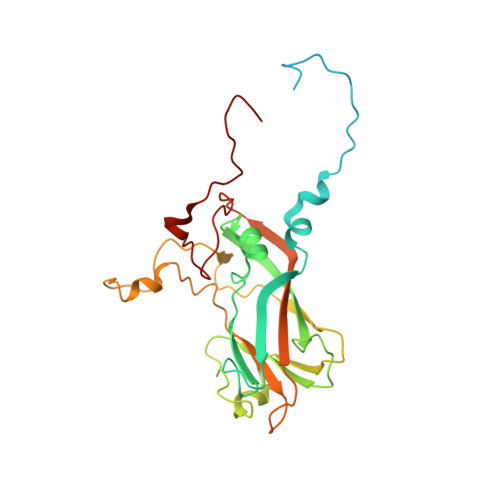
Mechanism of enterovirus VP0 maturation cleavage based on the structure of a stabilised assembly intermediate.
Kingston, N.J., Snowden, J.S., Grehan, K., Hall, P.K., Hietanen, E.V., Passchier, T.C., Polyak, S.J., Filman, D.J., Hogle, J.M., Rowlands, D.J., Stonehouse, N.J.(2024) PLoS Pathog 20: e1012511-e1012511
- PubMed: 39298524
- DOI: https://doi.org/10.1371/journal.ppat.1012511
- Primary Citation of Related Structures:
9F5S, 9F6A - PubMed Abstract:
Molecular details of genome packaging are little understood for the majority of viruses. In enteroviruses (EVs), cleavage of the structural protein VP0 into VP4 and VP2 is initiated by the incorporation of RNA into the assembling virion and is essential for infectivity. We have applied a combination of bioinformatic, molecular and structural approaches to generate the first high-resolution structure of an intermediate in the assembly pathway, termed a provirion, which contains RNA and intact VP0. We have demonstrated an essential role of VP0 E096 in VP0 cleavage independent of RNA encapsidation and generated a new model of capsid maturation, supported by bioinformatic analysis. This provides a molecular basis for RNA-dependence, where RNA induces conformational changes required for VP0 maturation, but that RNA packaging itself is not sufficient to induce maturation. These data have implications for understanding production of infectious virions and potential relevance for future vaccine and antiviral drug design.
- Astbury Centre for Structural Molecular Biology, School of Molecular & Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, United Kingdom.
Organizational Affiliation: