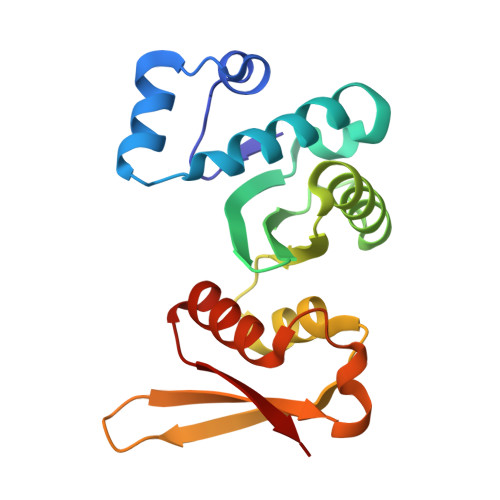
Archaeal NusA2 is the ancestor of ribosomal protein eS7 in eukaryotes.
Phung, D.K., Pilotto, S., Matelska, D., Blombach, F., Pinotsis, N., Hovan, L., Gervasio, F.L., Werner, F.(2025) Structure 33: 149
- PubMed: 39504966
- DOI: https://doi.org/10.1016/j.str.2024.10.019
- Primary Citation of Related Structures:
9F9U - PubMed Abstract:
N-utilization substance A (NusA) is a regulatory factor with pleiotropic functions in gene expression in bacteria. Archaea encode two conserved small proteins, NusA1 and NusA2, with domains orthologous to the two RNA binding K Homology (KH) domains of NusA. Here, we report the crystal structures of NusA2 from Sulfolobus acidocaldarius and Saccharolobus solfataricus obtained at 3.1 Å and 1.68 Å, respectively. NusA2 comprises an N-terminal zinc finger followed by two KH-like domains lacking the GXXG signature. Despite the loss of the GXXG motif, NusA2 binds single-stranded RNA. Mutations in the zinc finger domain compromise the structural integrity of NusA2 at high temperatures and molecular dynamics simulations indicate that zinc binding provides an energy barrier preventing the domain from reaching unfolded states. A structure-guided phylogenetic analysis of the KH-like domains supports the notion that the NusA2 clade is ancestral to the ribosomal protein eS7 in eukaryotes, implying a potential role of NusA2 in translation.
- RNAP Laboratory, Institute for Structural and Molecular Biology, University College London, London WC1E 6BT, UK.
Organizational Affiliation: