Structure of Trypanosoma peroxisomal import complex unveils conformational dynamics
Sonani, R.R., Blat, A., Jemiola-Rzeminska, M., Lipinski, O., Patel, S.N., Sood, T., Dubin, G.(2024) bioRxiv
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2024) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| malate dehydrogenase | 331 | Trypanosoma cruzi strain CL Brener | Mutation(s): 0 Gene Names: Tc00.1047053506503.69 EC: 1.1.1.37 | 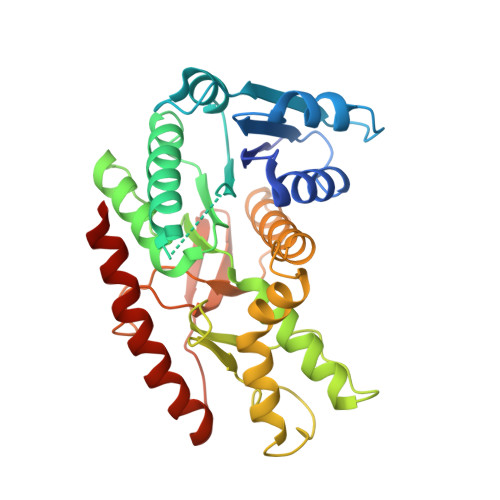 | |
UniProt | |||||
Find proteins for Q4DRD8 (Trypanosoma cruzi (strain CL Brener)) Explore Q4DRD8 Go to UniProtKB: Q4DRD8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q4DRD8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Polish National Science Centre | Poland | 2017/26/M/NZ1/00797 |
| Foundation for Polish Science | Poland | TEAM TECH CORE FACILITY/2017-4/6 |
| Polish National Science Centre | Poland | 2020/39/B/NZ1/01551 |