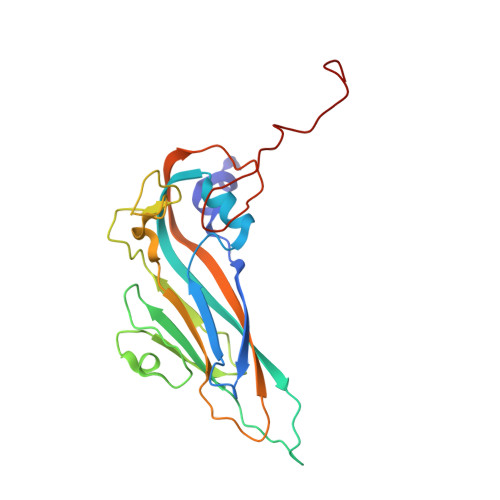
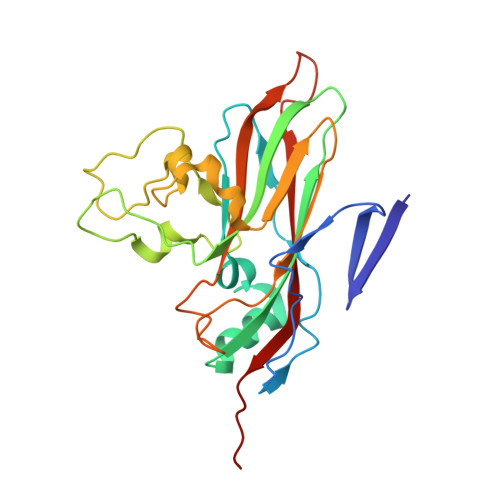
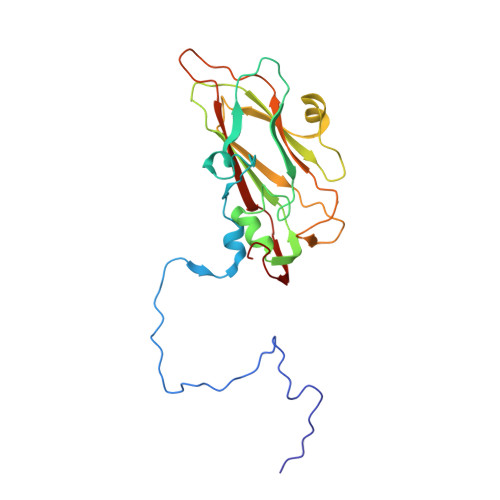
Comparison of structure and immunogenicity of CVB1-VLP and inactivated CVB1 vaccine candidates.
Soppela, S., Plavec, Z., Grohn, S., Jartti, M., Oikarinen, S., Laajala, M., Marjomaki, V., Butcher, S.J., Hankaniemi, M.M.(2024) Res Sq
- PubMed: 38978565
- DOI: https://doi.org/10.21203/rs.3.rs-4545395/v1
- Primary Citation of Related Structures:
9FJC, 9FJD, 9FJE - PubMed Abstract:
Coxsackievirus B1 (CVB1) is a common cause of acute and chronic myocarditis, dilated cardiomyopathy and aseptic meningitis. However, no CVB-vaccines are available for human use. In this study, we investigated the immunogenicity of virus-like particle (VLP) and inactivated whole-virus vaccines for CVB1 when administrated to mice via either subcutaneous or intranasal routes formulated with and without commercial and experimental adjuvants. Here, the potential of utilizing epigallocatechin-3-gallate (EGCG) as a mucosal adjuvant synergistically with its ability to inactivate the virus were investigated. EGCG had promising adjuvant properties for CVB1-VLP when administered via the parenteral route but limited efficacy via intranasal administration. However, intranasal administration of the formalin-inactivated virus induced high CVB1-specific humoral, cellular, and mucosal immune responses. Also, based on CVB1-specific IgG-antibody responses, we conclude that CVB1-VLP can be taken up by immune cells when administrated intranasally and further structural engineering for the VLP may increase the mucosal immunogenicity. The preparations contained mixtures of compact and expanded A particles with 85% expanded in the formalin-inactivated virus, but only 52% in the VLP observed by cryogenic electron microscopy. To correlate the structure to immunogenicity, we solved the structures of the CVB1-VLP and the formalin-inactivated CVB1 virus at resolutions ranging from 2.15 A to 4.1 A for the expanded and compact VLP and virus particles by image reconstruction. These structures can be used in designing mutations increasing the stability and immunogenicity of CVB1-VLP in the future. Overall, our results highlight the potential of using formalin inactivated CVB1 vaccine in mucosal immunization programs and provide important information for future development of VLP-based vaccines against all enteroviruses.
- Tampere University.
Organizational Affiliation: