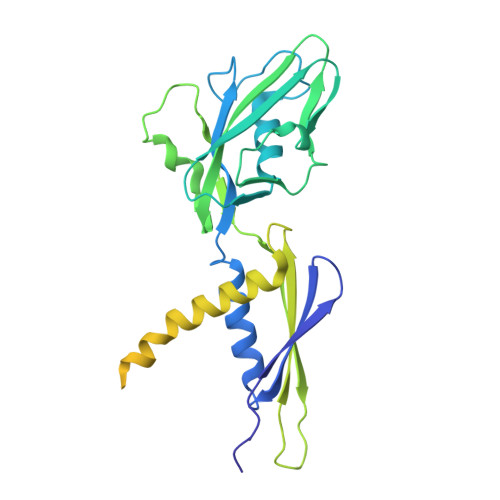
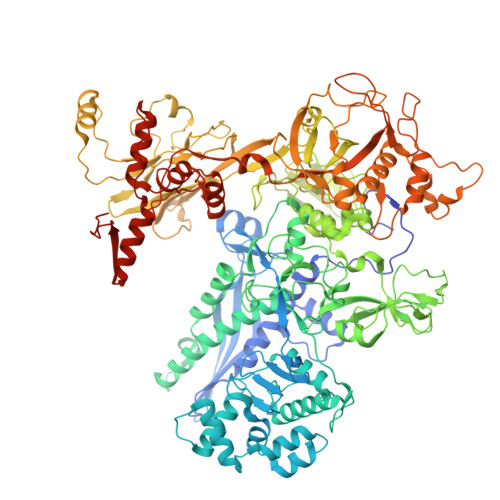
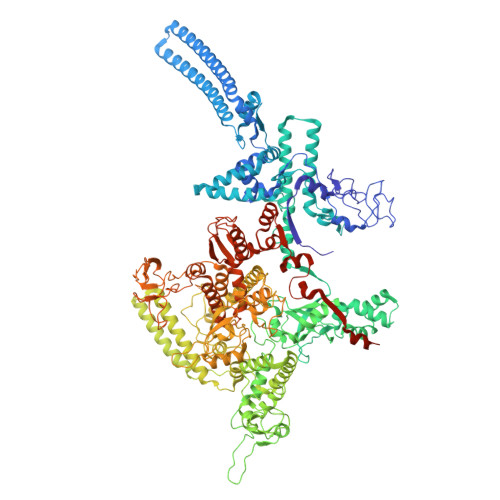
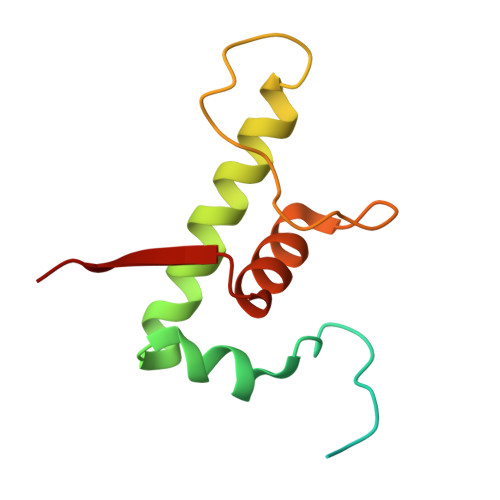
Single-stranded DNA drives sigma subunit loading onto mycobacterial RNA polymerase to unlock initiation-competent conformations.
Vishwakarma, R.K., Marechal, N., Morichaud, Z., Blaise, M., Margeat, E., Brodolin, K.(2025) Nucleic Acids Res 53
- PubMed: 40240004
- DOI: https://doi.org/10.1093/nar/gkaf272
- Primary Citation of Related Structures:
9FJP, 9FJR, 9FJS - PubMed Abstract:
Initiation of transcription requires the formation of the "open" promoter complex (RPo). For this, the σ subunit of bacterial RNA polymerase (RNAP) binds to the nontemplate strand of the -10 element sequence of promoters and nucleates DNA unwinding. This is accompanied by a cascade of conformational changes on RNAP, the exact mechanics of which remains elusive. Here, using single-molecule Förster resonance energy transfer and cryo-electron microscopy, we explored the conformational landscape of RNAP from the human pathogen Mycobacterium tuberculosis upon binding to a single-stranded DNA (ssDNA) fragment that includes the -10 element sequence (-10 ssDNA). We found that like the transcription activator RNAP-binding protein A, -10 ssDNA induced σ subunit loading onto the DNA/RNA channels of RNAP. This triggered RNAP clamp closure and unswiveling that are required for RPo formation and RNA synthesis initiation. Our results reveal a mechanism of ssDNA-guided RNAP maturation and identify the σ subunit as a regulator of RNAP conformational dynamics.
- Institut de Recherche en Infectiologie de Montpellier, Univ Montpellier, CNRS, Montpellier 34293, France.
Organizational Affiliation: