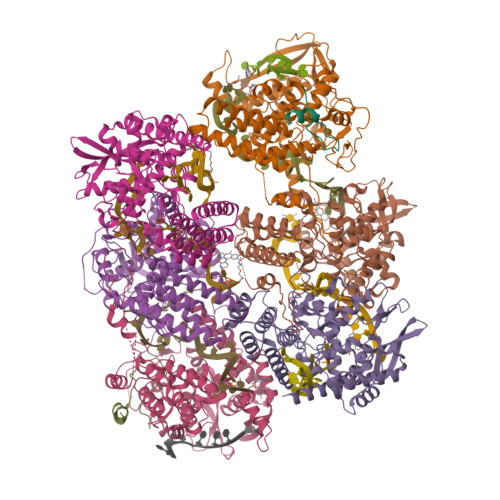
Influenza a virus antiparallel helical nucleocapsid-like pseudo-atomic structure.
Chenavier, F., Zarkadas, E., Freslon, L.L., Stelfox, A.J., Schoehn, G., Ruigrok, R.W.H., Ballandras-Colas, A., Crepin, T.(2025) Nucleic Acids Res 53
- PubMed: 39673795
- DOI: https://doi.org/10.1093/nar/gkae1211
- Primary Citation of Related Structures:
9GAN, 9GAP, 9GAQ, 9GAS, 9GAT, 9GAV - PubMed Abstract:
Influenza A viruses are responsible for human seasonal epidemics and severe animal pandemics with a risk of zoonotic transmission to humans. The viral segmented RNA genome is encapsidated by nucleoproteins (NP) and attached to the heterotrimeric polymerase, forming the viral ribonucleoproteins (vRNPs). Flexible helical vRNPs are central for viral transcription and replication. In this study, we present an advanced biological tool, the antiparallel helical RNP-like complex, assembled from recombinant N-terminally truncated NP and short synthetic RNA. The 3.0 Å cryo-electron microscopy structure details for the first time the whole RNA pathway across NP as well as NP-NP interactions that drive the antiparallel helical assembly accommodating major and minor grooves. Our findings show that the surface of the protein can harbour several conformations of the RNA, confirming that the number of nucleobases that binds to NP is not fixed, but ranges probably between 20 and 24. Taking all together, our data provide details to further understand the genome encapsidation and explain the inherent flexibility of influenza A virus vRNPs.
Organizational Affiliation:
Univ. Grenoble Alpes, CNRS, CEA, IBS, 71 avenue des Martyrs, F-38000 Grenoble, France.