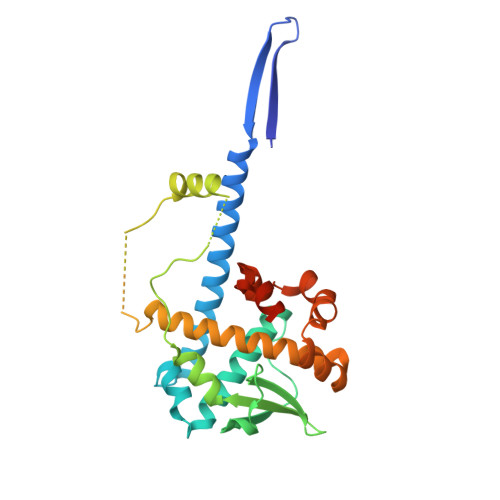
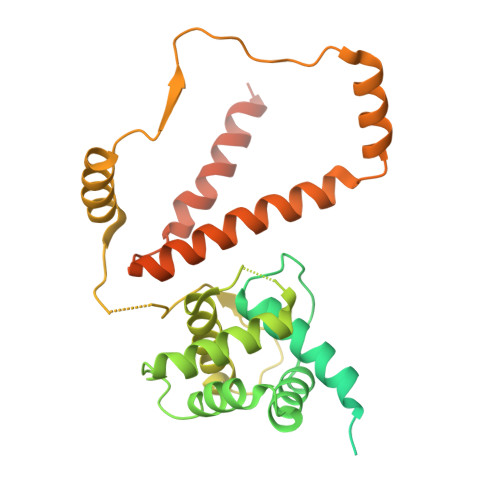
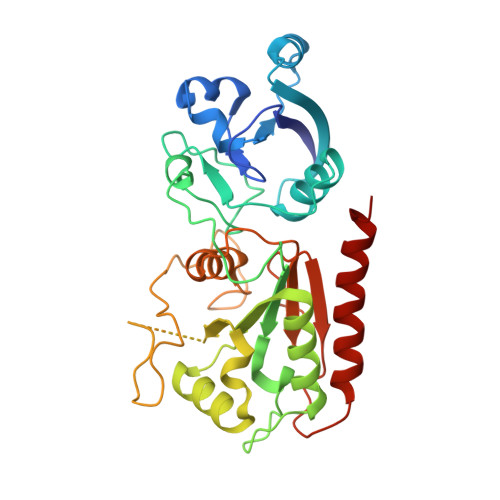
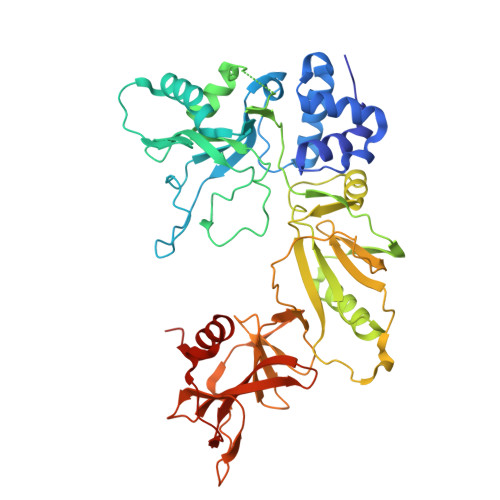
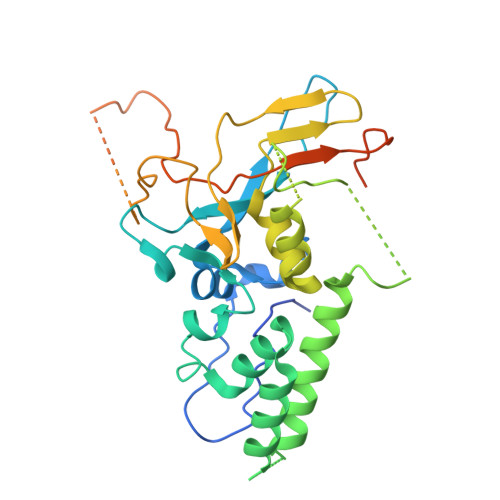
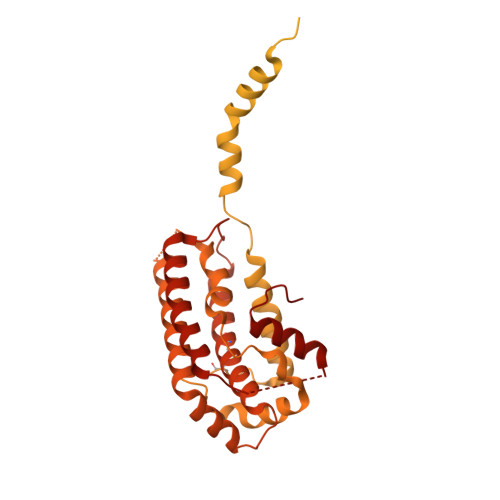
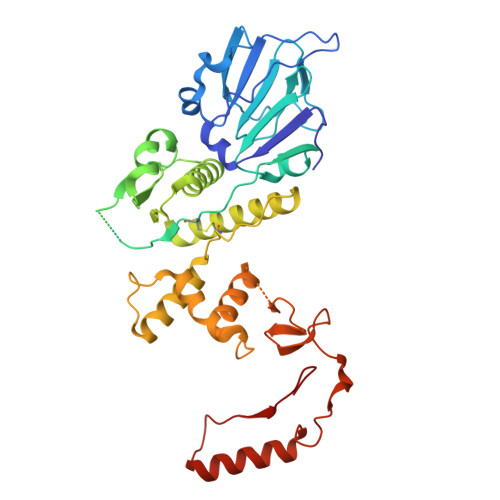
Structure of AcMNPV nucleocapsid reveals DNA portal organization and packaging apparatus of circular dsDNA baculovirus.
Effantin, G., Kandiah, E., Pelosse, M.(2025) Nat Commun 16: 4844-4844
- PubMed: 40413174
- DOI: https://doi.org/10.1038/s41467-025-60152-2
- Primary Citation of Related Structures:
9H1S, 9H2A, 9H2B, 9H2C, 9H2H, 9H2J, 9H2K - PubMed Abstract:
Baculoviruses are large DNA viruses found in nature propagating amongst insects and lepidoptera in particular. They have been studied for decades and are nowadays considered as invaluable biotechnology tools used as biopesticides, recombinant expression systems or delivery vehicle for gene therapy. However, little is known about the baculovirus nucleocapsid assembly at a molecular level. Here, we solve the whole structure of the Autographa californica multiple nucleopolyhedrovirus (AcMNPV) nucleocapsid by applying cryo-electron microscopy (CryoEM) combined with de novo modelling and Alphafold predictions. Our structure completes prior observations and elucidates the intricate architecture of the apical cap, unravelling the organization of a DNA portal featuring intriguing symmetry mismatches between its core and vertex. The core, closing the capsid at the apex, holds two DNA helices of the viral genome tethered to Ac54 proteins. Different symmetry components at the apical cap and basal structure are constituted of the same building block, made of Ac101/Ac144, proving the versatility of this modular pair. The crown forming the portal vertex displays a C21 symmetry and contains, amongst others, the motor-like protein Ac66. Our findings support the viral portal to be involved in DNA packaging, probably in conjunction with other parts of a larger DNA packaging apparatus.
Organizational Affiliation:
Univ. Grenoble Alpes, CNRS, CEA, Institut de Biologie Structurale (IBS), 38000, Grenoble, France. gregory.effantin@ibs.fr.