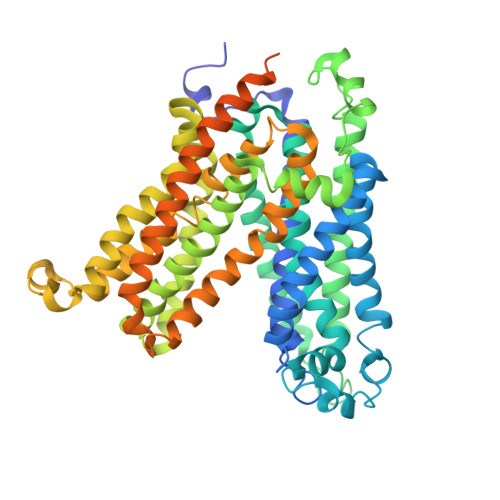
Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Lin, H., Huang, J., Li, T., Li, W., Wu, Y., Yang, T., Nian, Y., Lin, X., Wang, J., Wang, R., Zhao, X., Su, N., Zhang, J., Wu, X., Fan, M.(2025) Nature 641: 797-804
- PubMed: 40074904
- DOI: https://doi.org/10.1038/s41586-025-08743-3
- Primary Citation of Related Structures:
9J3J, 9J3L, 9J3M, 9J3N, 9J3O - PubMed Abstract:
Adenosine triphosphate (ATP) is the principal energy currency of all living cells 1,2 . Metabolically impaired obligate intracellular parasites, such as the human pathogens Chlamydia trachomatis and Rickettsia prowazekii, can acquire ATP from their host cells through a unique ATP/adenosine diphosphate (ADP) translocator, which mediates the import of ATP into and the export of ADP and phosphate out of the parasite cells, thus allowing the exploitation of the energy reserves of host cells (also known as energy parasitism). This type of ATP/ADP translocator also exists in the obligate intracellular endosymbionts of protists and the plastids of plants and algae and has been implicated to play an important role in endosymbiosis 3-31 . The plastid/parasite type of ATP/ADP translocator is phylogenetically and functionally distinct from the mitochondrial ATP/ADP translocator, and its structure and transport mechanism are still unknown. Here we report the cryo-electron microscopy structures of two plastid/parasite types of ATP/ADP translocators in the apo and substrate-bound states. The ATP/ADP-binding pocket is located at the interface between the N and C domains of the translocator, and a conserved asparagine residue within the pocket is critical for substrate specificity. The translocator operates through a rocker-switch alternating access mechanism involving the relative rotation of the two domains as rigid bodies. Our results provide critical insights for understanding ATP translocation across membranes in energy parasitism and endosymbiosis and offer a structural basis for developing drugs against obligate intracellular parasites.
- CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: