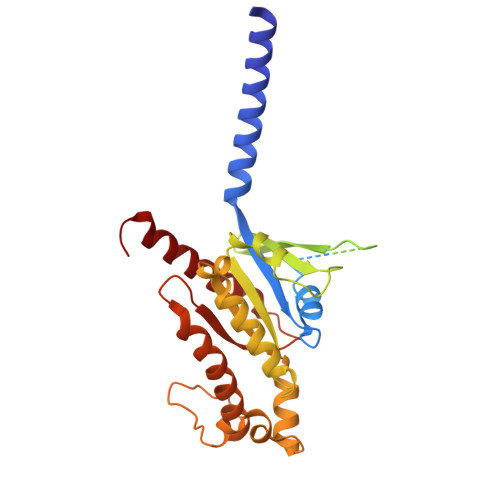
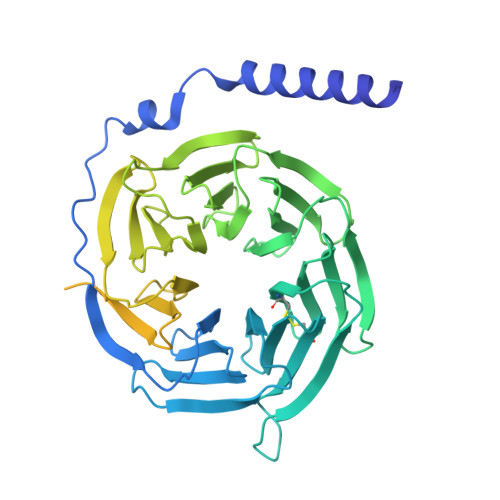
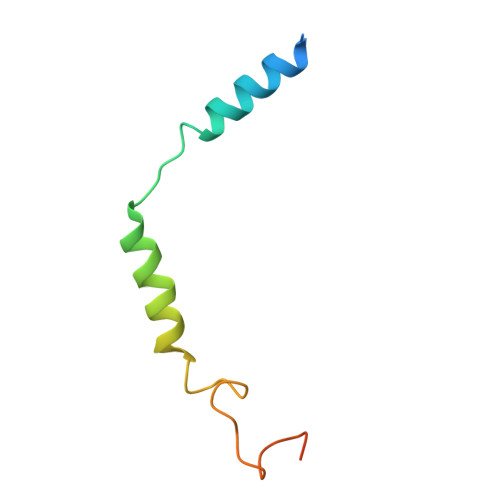
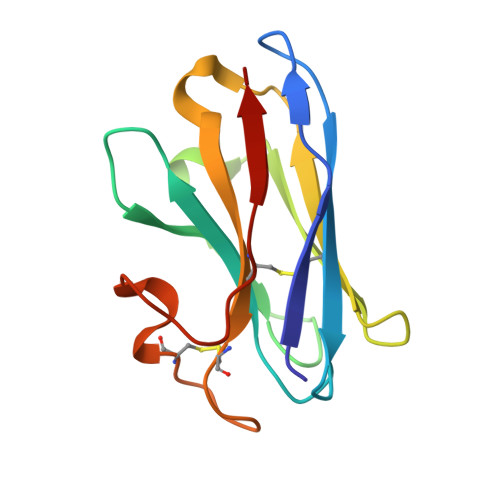
Molecular mechanism of pH sensing and activation in GPR4 reveals proton-mediated GPCR signaling.
You, C., Guo, S., Zhang, T., He, X., Gao, T., Xin, W., Zhu, Z., Lu, Y., Xu, Y., Li, Z., Zhang, Y., Cheng, X., Jiang, Y., Xie, X., Xu, H.E.(2025) Cell Discov 11: 59-59
- PubMed: 40555728
- DOI: https://doi.org/10.1038/s41421-025-00807-y
- Primary Citation of Related Structures:
9JCO, 9JCP, 9JCQ - PubMed Abstract:
Maintaining pH homeostasis is critical for cellular function across all living organisms. Proton-sensing G protein-coupled receptors (GPCRs), particularly GPR4, play a pivotal role in cellular responses to pH changes. Yet, the molecular mechanisms underlying their proton sensing and activation remain incompletely understood. Here we present high-resolution cryo-electron microscopy structures of GPR4 in complex with G proteins under physiological and acidic pH conditions. Our structures reveal an intricate proton-sensing mechanism driven by a sophisticated histidine network in the receptor's extracellular domain. Upon protonation of key histidines under acidic conditions, a remarkable conformational cascade is initiated, propagating from the extracellular region to the intracellular G protein-coupling interface. This dynamic process involves precise transmembrane helix rearrangements and conformational shifts of conserved motifs, mediated by strategically positioned water molecules. Notably, we discovered a bound bioactive lipid, lysophosphatidylcholine, which has positive allosteric effects on GPR4 activation. These findings provide a comprehensive framework for understanding proton sensing in GPCRs and the interplay between pH sensing and lipid regulation, offering insights into cellular pH homeostasis and potential therapies for pH-related disorders.
Organizational Affiliation:
The State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China. youchongzhao@gmail.com.